Contents
Plot figures from experimental dataset for the publication:
Rindal, O. M. H., Rodriguez-Molares, A., & Austeng, A. (2017). The Dark Region Artifact in Adaptive Ultrasound Beamforming. IEEE International Ultrasonics Symposium, IUS, 1-4.
clear all;close all % data location url='http://ustb.no/datasets/'; % if not found downloaded from here local_path = [ustb_path(),'/data/']; % location of example data % Choose dataset filename='beamformed_experimental_data.uff'; % check if the file is available in the local path or downloads otherwise tools.download(filename, url, local_path); % Read the beamformed images from the dataset b_data_das = uff.read_object([local_path filename],'/b_data_das'); b_data_mv = uff.read_object([local_path filename],'/b_data_mv'); b_data_ebmv = uff.read_object([local_path filename],'/b_data_ebmv'); b_data_cf = uff.read_object([local_path filename],'/b_data_cf'); b_data_gcf = uff.read_object([local_path filename],'/b_data_gcf'); b_data_pcf = uff.read_object([local_path filename],'/b_data_pcf'); b_data_dmas = uff.read_object([local_path filename],'/b_data_dmas');
UFF: reading b_data_das [uff.beamformed_data] UFF: reading b_data_mv [uff.beamformed_data] UFF: reading b_data_ebmv [uff.beamformed_data] UFF: reading b_data_cf [uff.beamformed_data] UFF: reading b_data_gcf [uff.beamformed_data] UFF: reading b_data_pcf [uff.beamformed_data] UFF: reading b_data_dmas [uff.beamformed_data]
Let's write out some info about the authorship of this dataset
b_data_das.print_authorship
Name: Beamformed dataset used to create the figures in the publications under reference. Reference: Rindal, O. M. H., Rodriguez-Molares, A., & Austeng, A. (2017). The Dark Region Artifact in Adaptive Ultrasound Beamforming. IEEE International Ultrasonics Symposium, IUS, 1-4. Author(s): Ole Marius Hoel Rindal <olemarius@olemarius.net> Version: 1.0.0
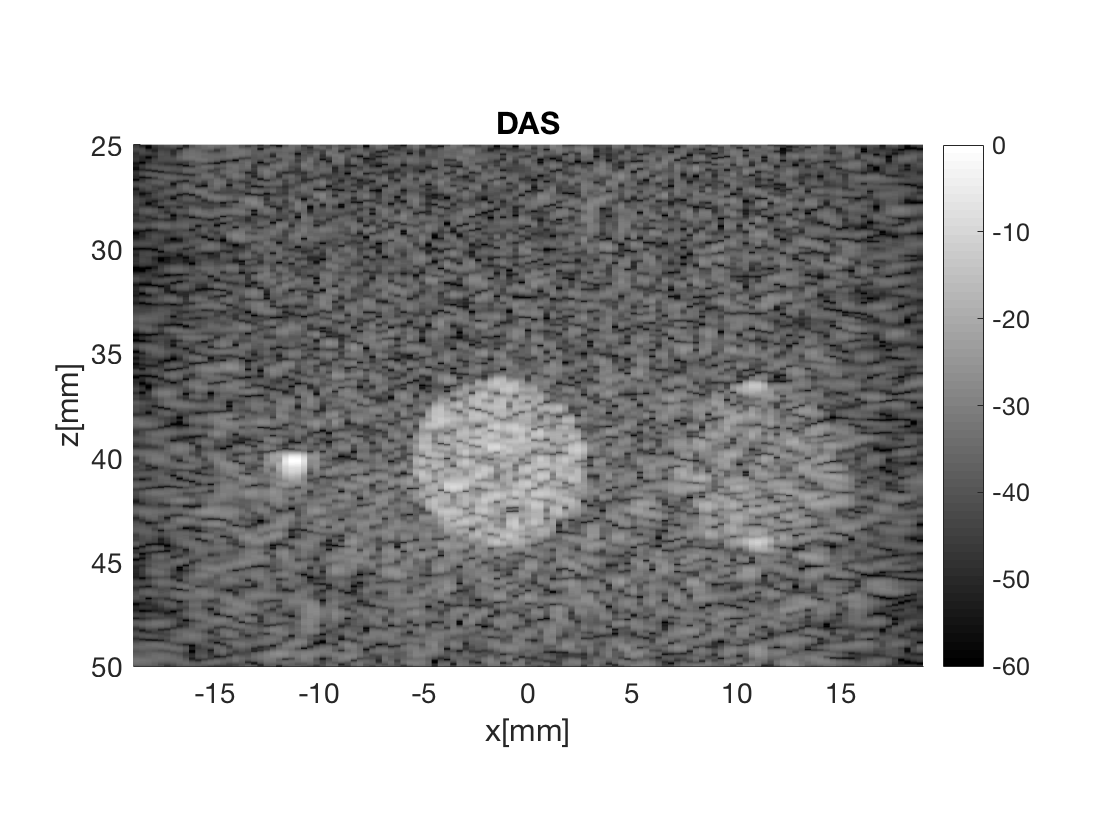
Display the images in Fig. 3
Massage the images into a cell array for easier processing in helper functions. Also display all images used in Fig. 3 in the publication.
image.all{1} = b_data_das.get_image();
image.tags{1} = 'DAS';
b_data_das.plot(1,'DAS');
image.all{2} = b_data_mv.get_image();
image.tags{2} = 'MV';
b_data_mv.plot(2,'MV');
image.all{3} = b_data_ebmv.get_image();
image.tags{3} = 'EBMV';
b_data_ebmv.plot(3,'EBMV');
image.all{4} = b_data_cf.get_image();
image.tags{4} = 'CF';
b_data_cf.plot(4,'CF');
image.all{5} = b_data_gcf.get_image();
image.tags{5} = 'GCF';
b_data_gcf.plot(5,'GCF');
image.all{6} = b_data_pcf.get_image();
image.tags{6} = 'PCF';
b_data_pcf.plot(6,'PCF');
image.all{7} = b_data_dmas.get_image();
image.tags{7} = 'DMAS';
b_data_dmas.plot(7,'DMAS');
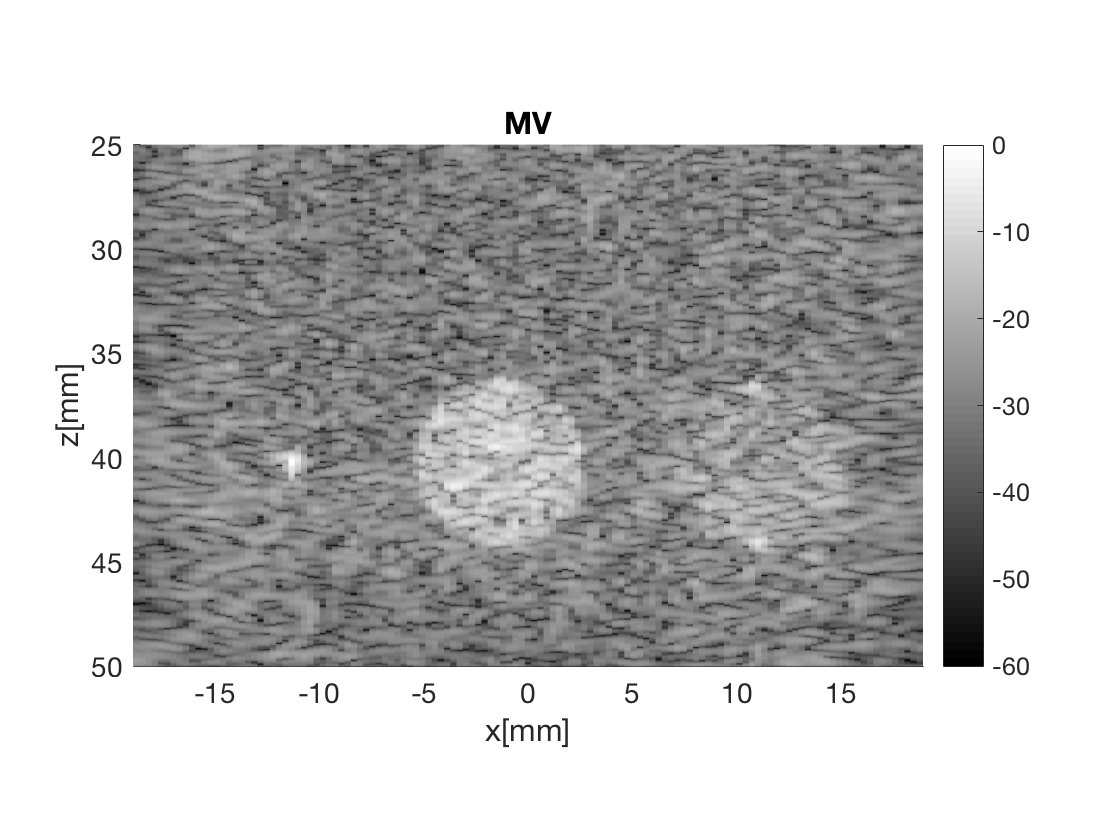
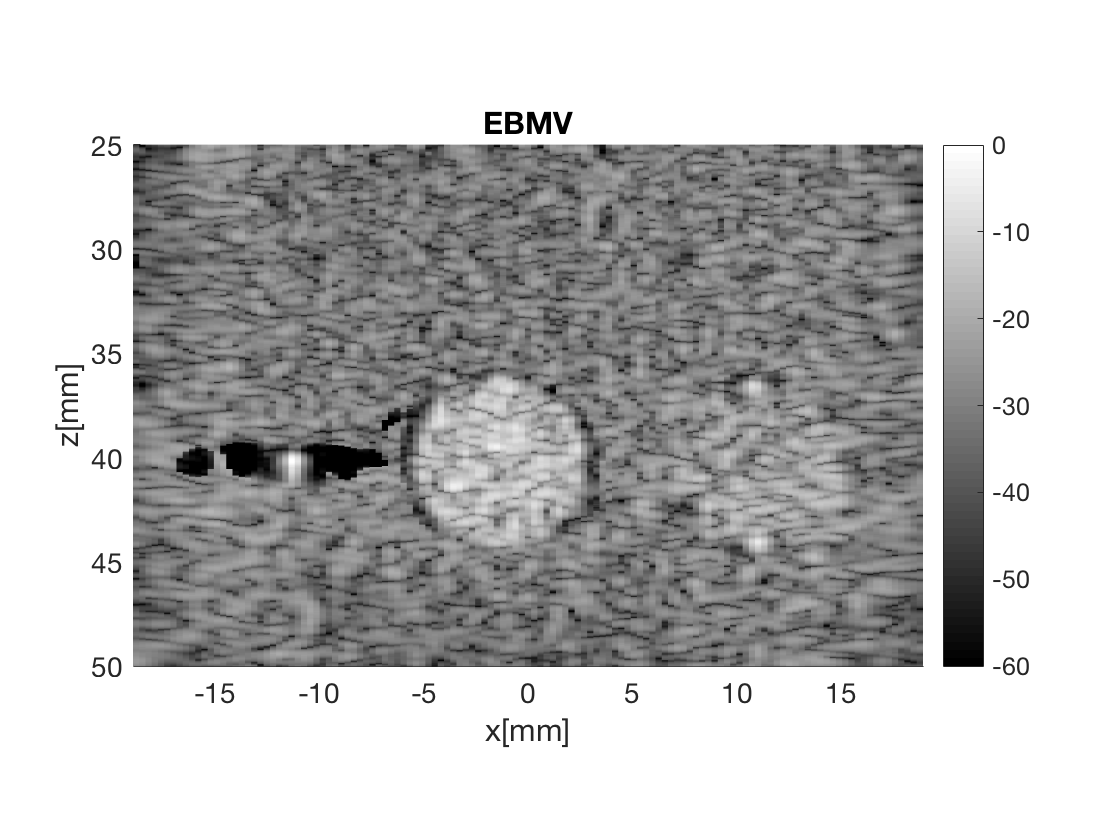
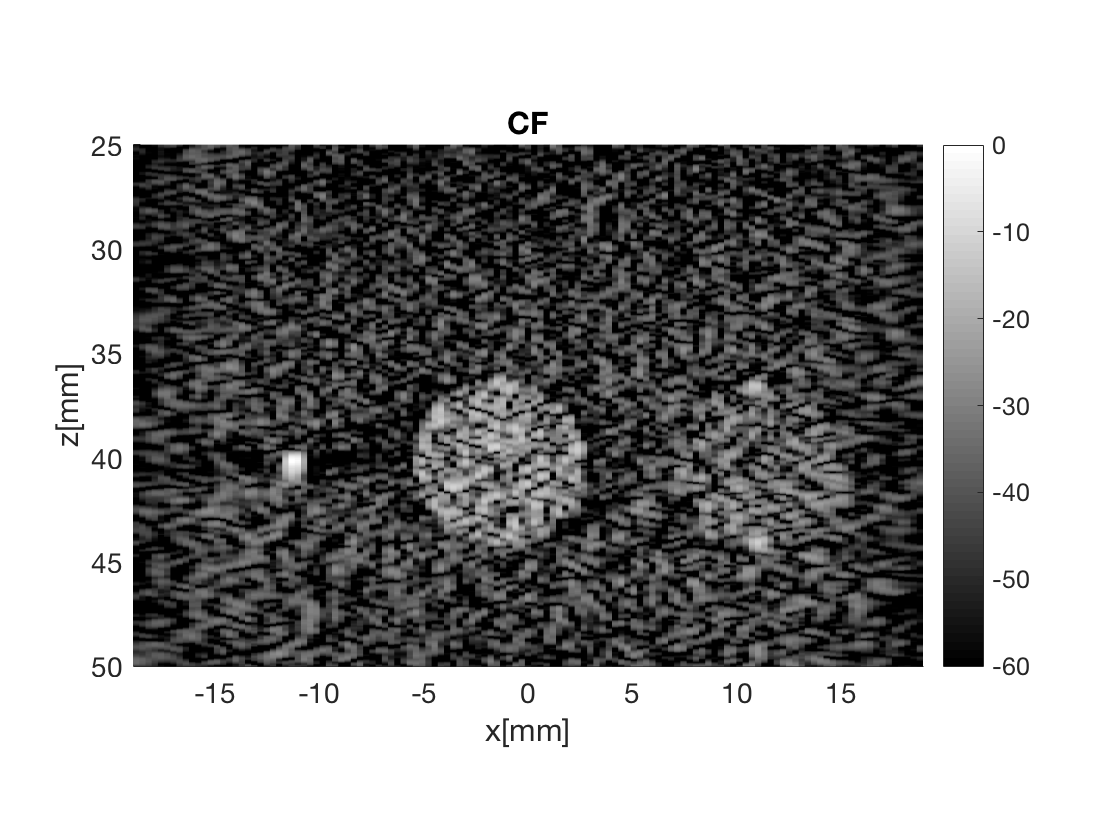
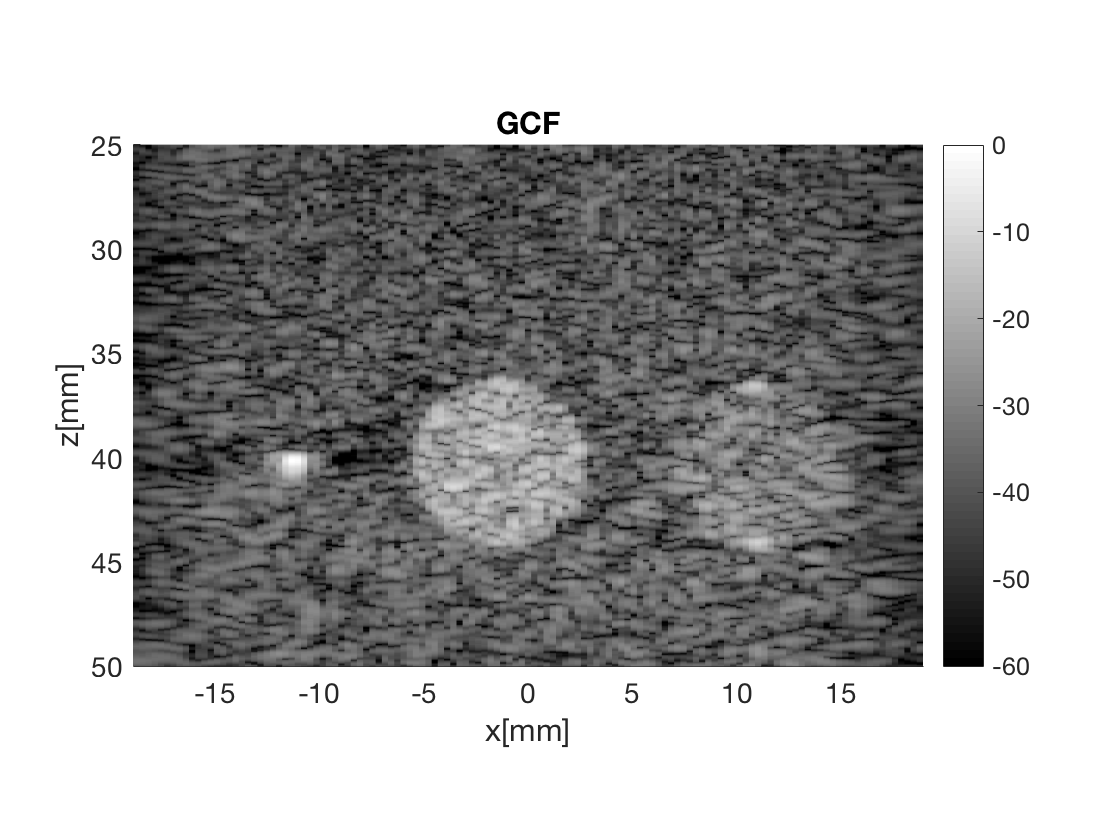
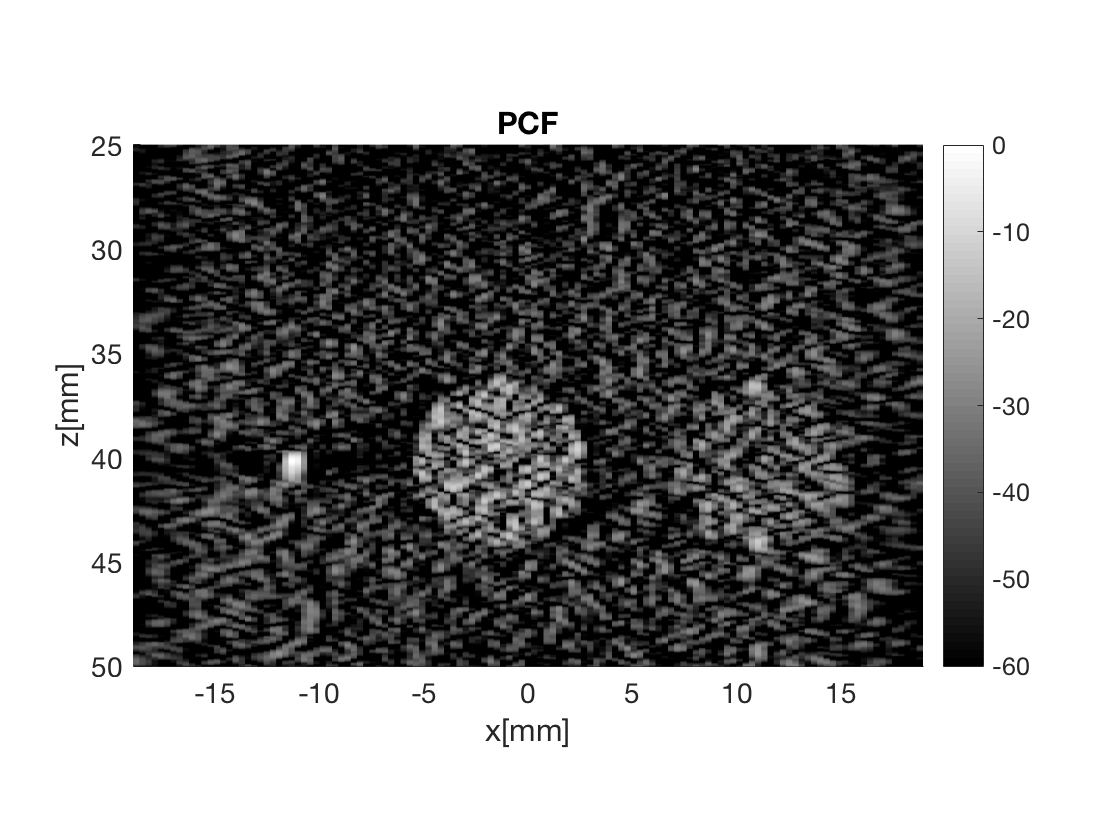
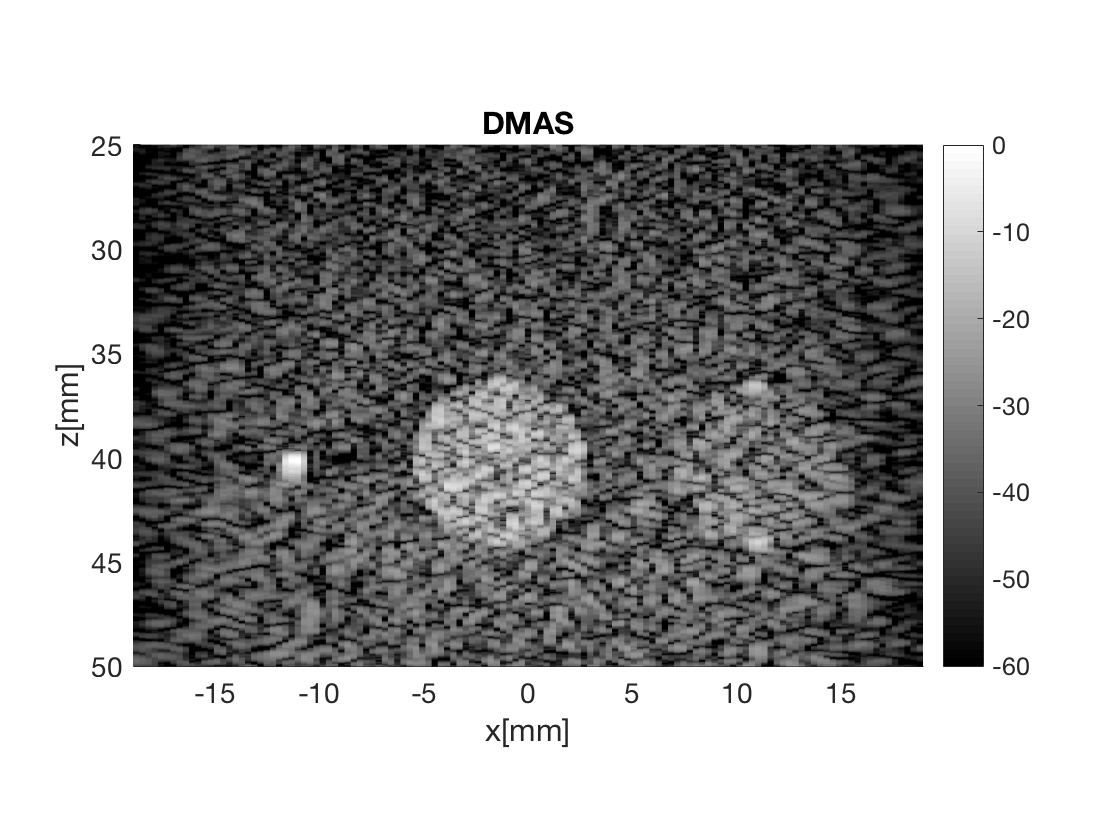
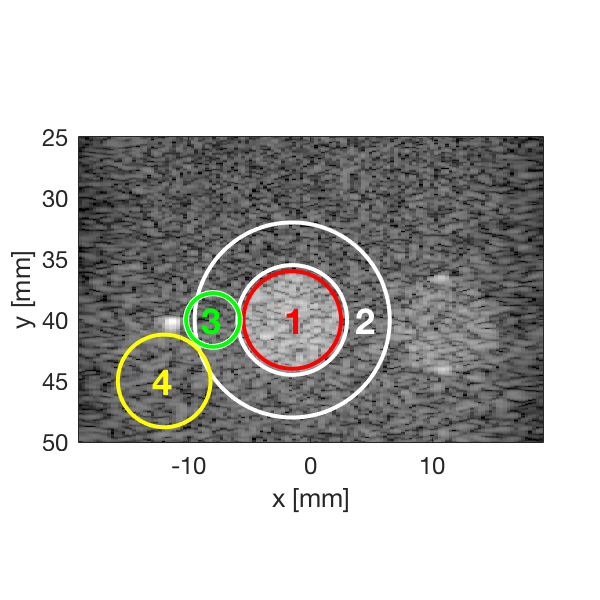
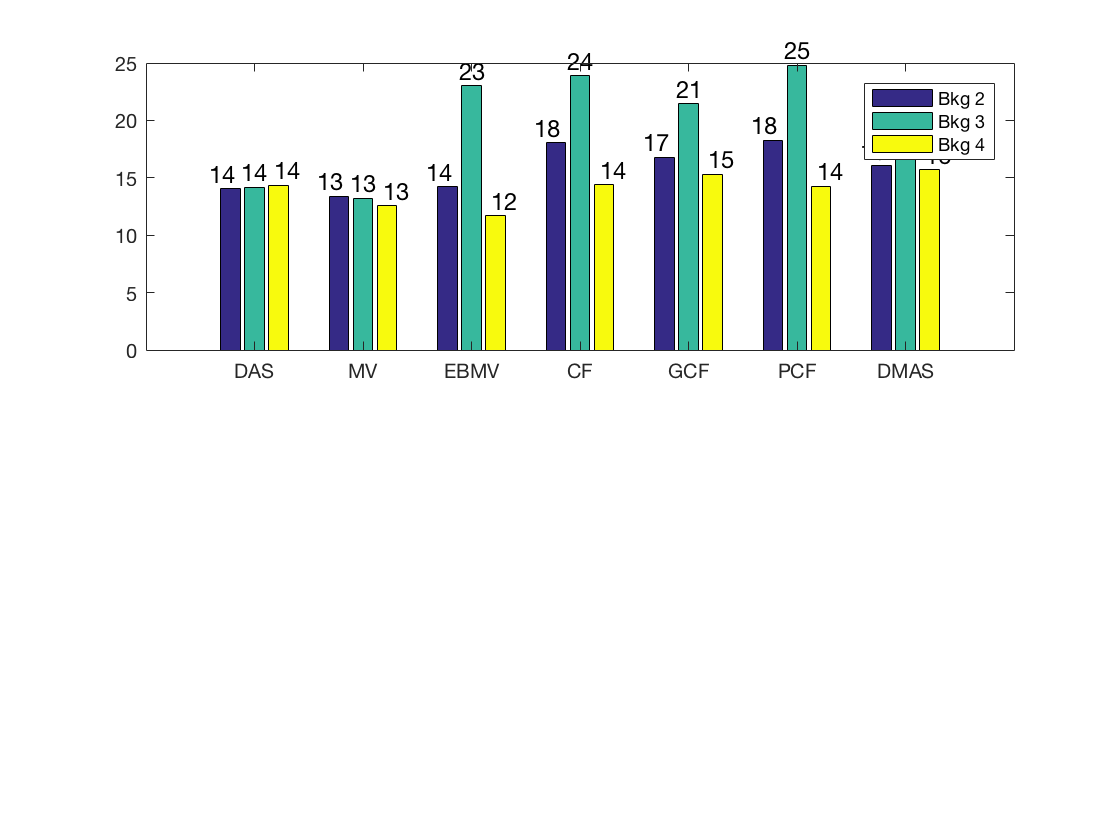
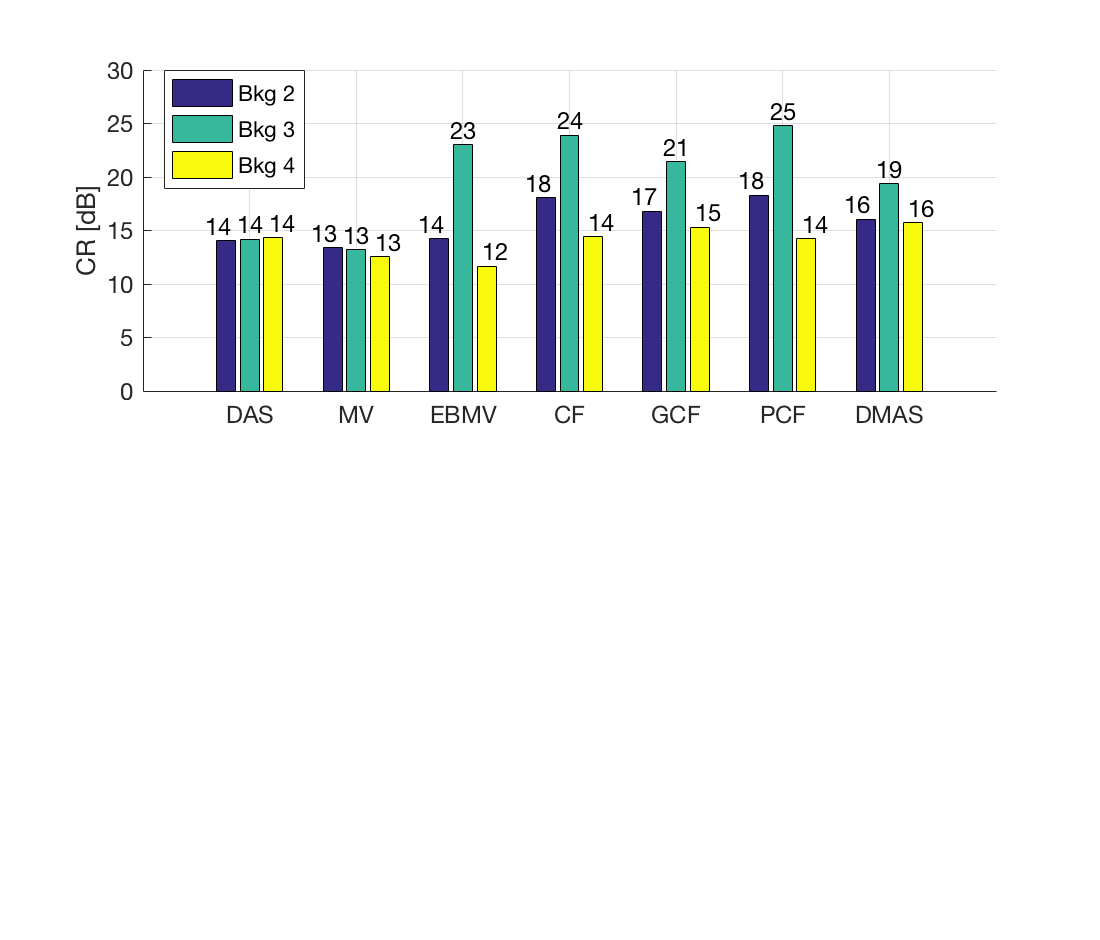
Measure the contrast creating Fig. 4
Measure the contrast of the different images creating Fig. 4 in the publication with the function measure_contrast_experimental_dra, also create the DAS image for Fig. 3 with regions indicated.
[CR,CR_alt,CR_alt_2,handle_DAS] = measure_contrast_experimental_dra... (b_data_das,image,-1.5,40,4,4.5,8,-12,45,2.2,-8,40,3.8); % Create bar plot for Fig. 4 in the publication. [handle] = plot_contrast_differences_DRA(CR,CR_alt_2,CR_alt,image,'Bkg 2','Bkg 3','Bkg 4'); f12= figure(12);clf set(f12,'Position',[100, 100, 550, 470]); ax = subplot(2,1,1); copyobj(allchild(handle),ax) set(gca,'XTick',linspace(1,length(image.tags),length(image.tags))) set(gca,'XTickLabel',image.tags) ylabel('CR [dB]'); h_legend = legend('show','Location','best'); grid on ylim([0 30]); set(gca,'FontSize',12); set(h_legend,'Position',[0.1525 0.7997 0.1209 0.1257]);