Contents
Run script to create beampatterns
create_beampatterns_plot
Load the data
clear all;
close all;
url='http://ustb.no/datasets/';
local_path = [data_path(), filesep];
filename = ['FieldII_CPWC_point_scatterers_res_v2.uff'];
tools.download(filename, url, local_path);
b_data_das = uff.beamformed_data();
b_data_cf = uff.beamformed_data();
b_data_pcf = uff.beamformed_data();
b_data_gcf = uff.beamformed_data();
b_data_mv = uff.beamformed_data();
b_data_ebmv = uff.beamformed_data();
b_data_dmas = uff.beamformed_data();
b_data_das.read([local_path,filesep,filename],'/b_data_das');
b_data_cf.read([local_path,filesep,filename],'/b_data_cf');
b_data_pcf.read([local_path,filesep,filename],'/b_data_pcf');
b_data_gcf.read([local_path,filesep,filename],'/b_data_gcf');
b_data_mv.read([local_path,filesep,filename],'/b_data_mv');
b_data_ebmv.read([local_path,filesep,filename],'/b_data_ebmv');
b_data_dmas.read([local_path,filesep,filename],'/b_data_dmas');
sca = b_data_das.scan;
separating_distance = [4 2 1 0.5 0.44 0.4];
UFF: reading /b_data_das [uff.beamformed_data]
UFF: reading /b_data_cf [uff.beamformed_data]
UFF: reading /b_data_pcf [uff.beamformed_data]
UFF: reading /b_data_gcf [uff.beamformed_data]
UFF: reading /b_data_mv [uff.beamformed_data]
UFF: reading /b_data_ebmv [uff.beamformed_data]
UFF: reading /b_data_dmas [uff.beamformed_data]
Plot the images
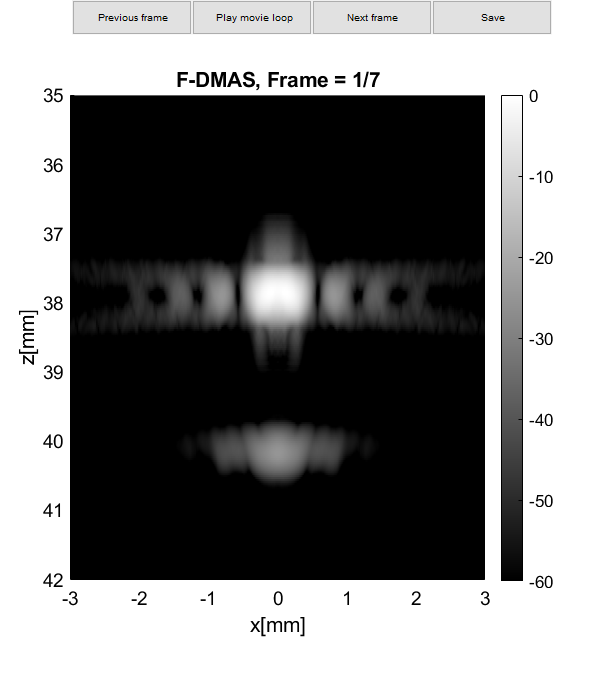
b_data_das.plot([],'DAS');
b_data_cf.plot([],'CF');
b_data_pcf.plot([],'PCF');
b_data_gcf.plot([],'GCF');
b_data_mv.plot([],'MV');
b_data_ebmv.plot([],'EBMV');
b_data_dmas.plot([],'F-DMAS');
img{1} = b_data_das.get_image();
img{2} = b_data_cf.get_image();
img{3} = b_data_pcf.get_image();
img{4} = b_data_gcf.get_image();
img{5} = b_data_mv.get_image();
img{6} = b_data_ebmv.get_image();
img{7} = b_data_dmas.get_image();
tags{1} = 'DAS';
tags{2} = 'CF';
tags{3} = 'PCF';
tags{4} = 'GCF';
tags{5} = 'MV';
tags{6} = 'EBMV';
tags{7} = 'F-DMAS';
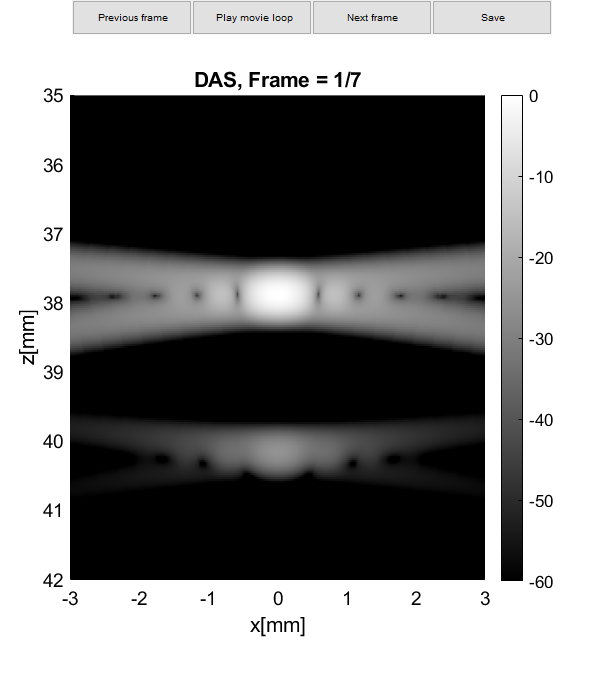
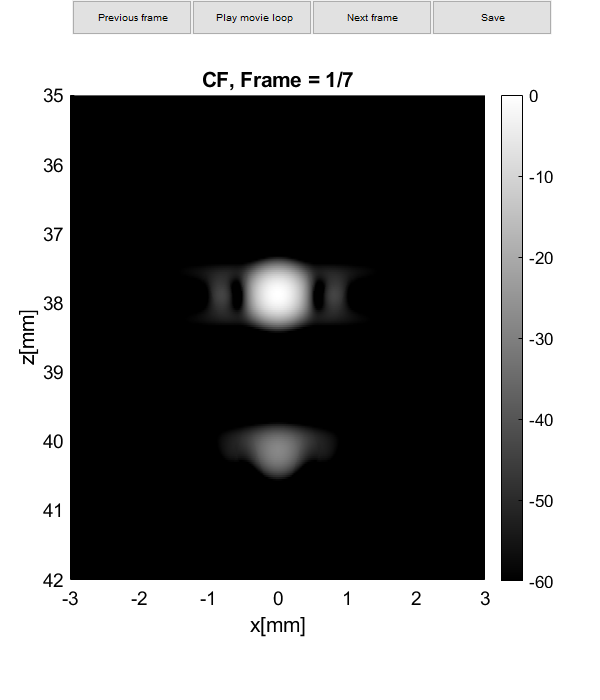
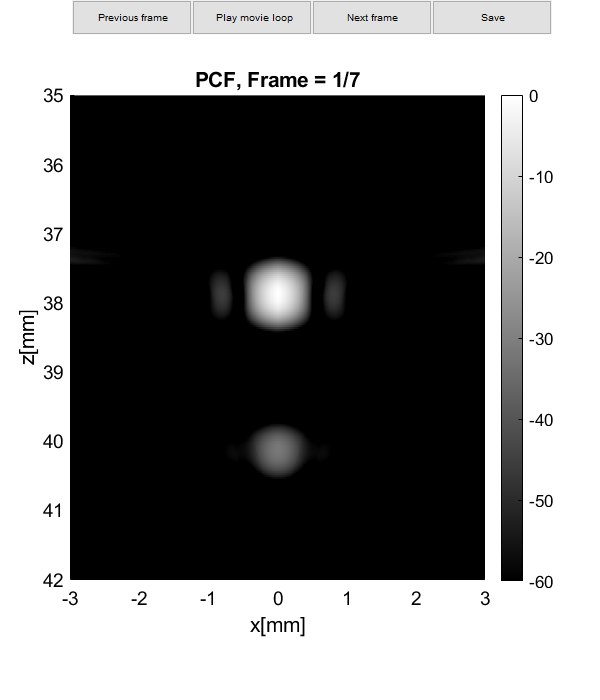
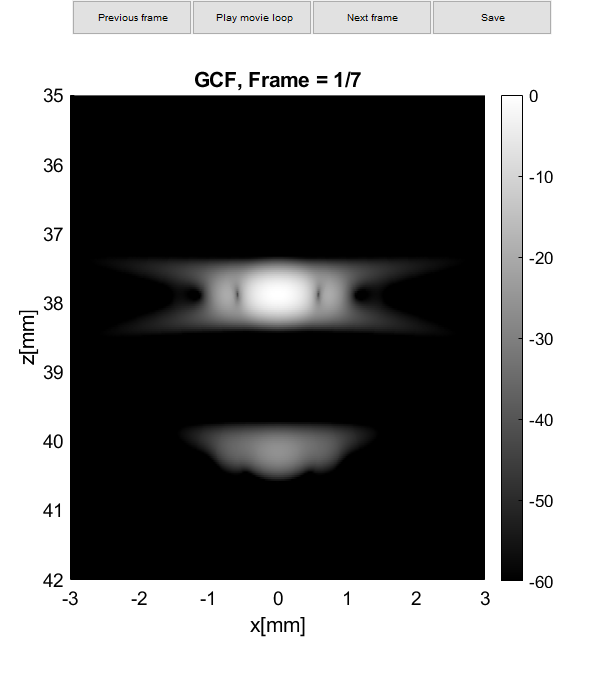
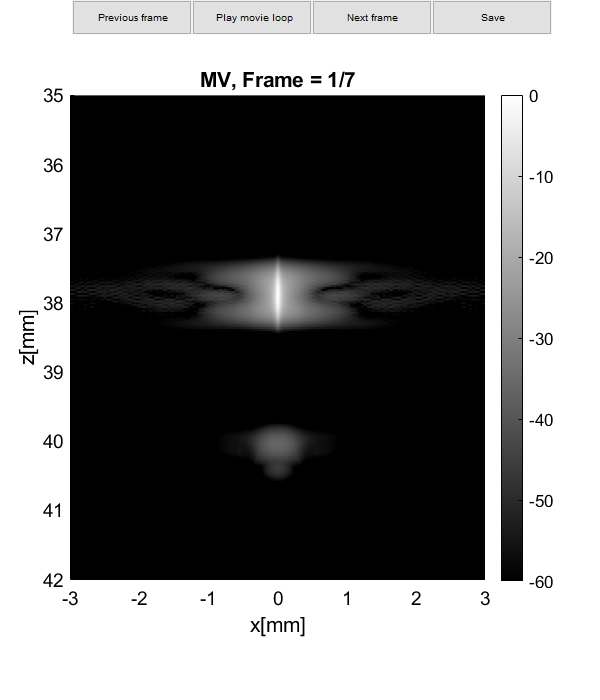
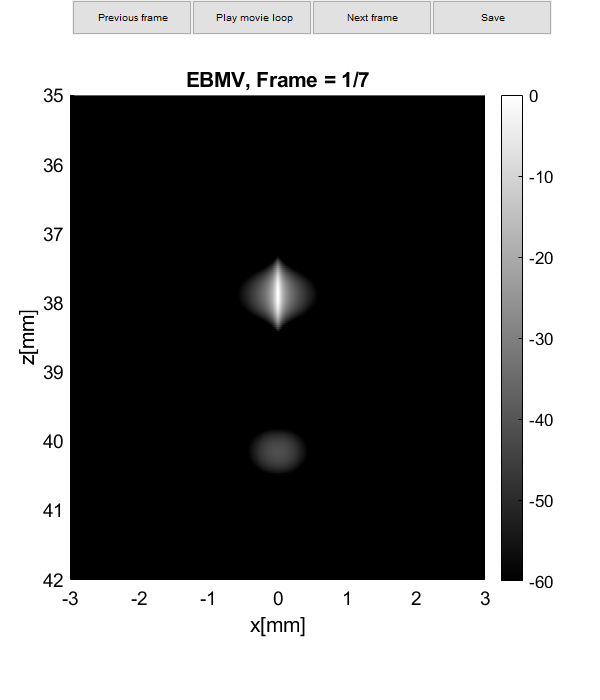
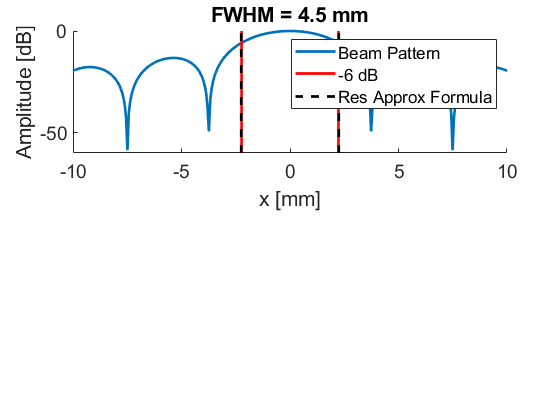
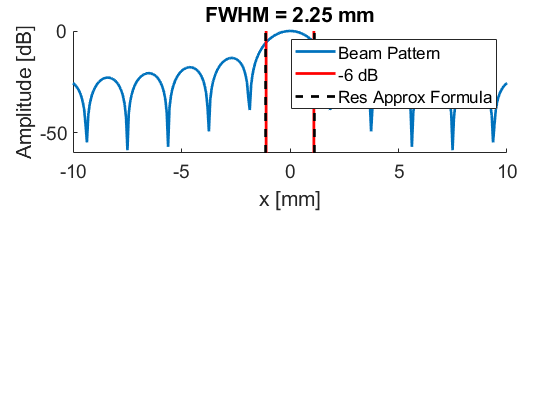
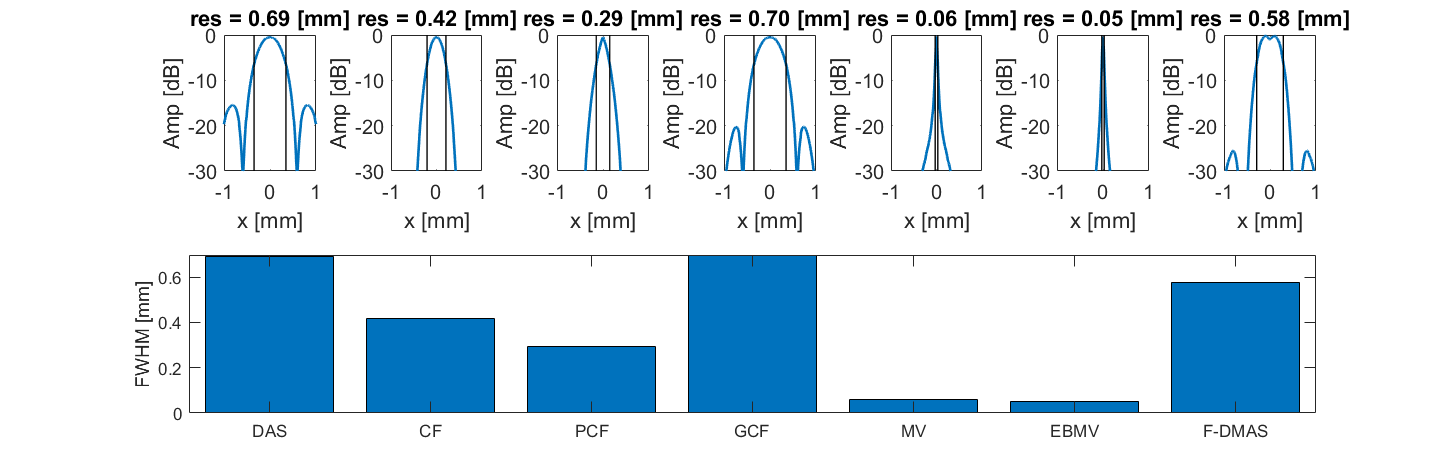
Plot resolution measured as FWHM
img_none{1} = b_data_das.get_image('none');
img_none{2} = b_data_cf.get_image('none');
img_none{3} = b_data_pcf.get_image('none');
img_none{4} = b_data_gcf.get_image('none');
img_none{5} = b_data_mv.get_image('none');
img_none{6} = b_data_ebmv.get_image('none');
img_none{7} = b_data_dmas.get_image('none');
f = figure(88);clf;
clear res;
for m = 1:7
format long
lateral = img{m}(103,:,1);
[res(m)] = calculate_6dB_resolution(sca.x_axis*10^3,lateral,1,88,m);
ylim(gca,[-30 0])
xlim(gca,[-1 1])
set(gca,'FontSize',15);
end
subplot(2,7,[8:14]);
bar(1:7,res)
xlim([0.5 7.5])
ylabel('FWHM [mm]');
xticks(1:7)
xticklabels(tags)
set(gca,'FontSize',13);
set(gcf,'Position',[10 163 1454 464])
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'FWHM_res_v2'],'eps2c')
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'FWHM_res_v2'],'png')
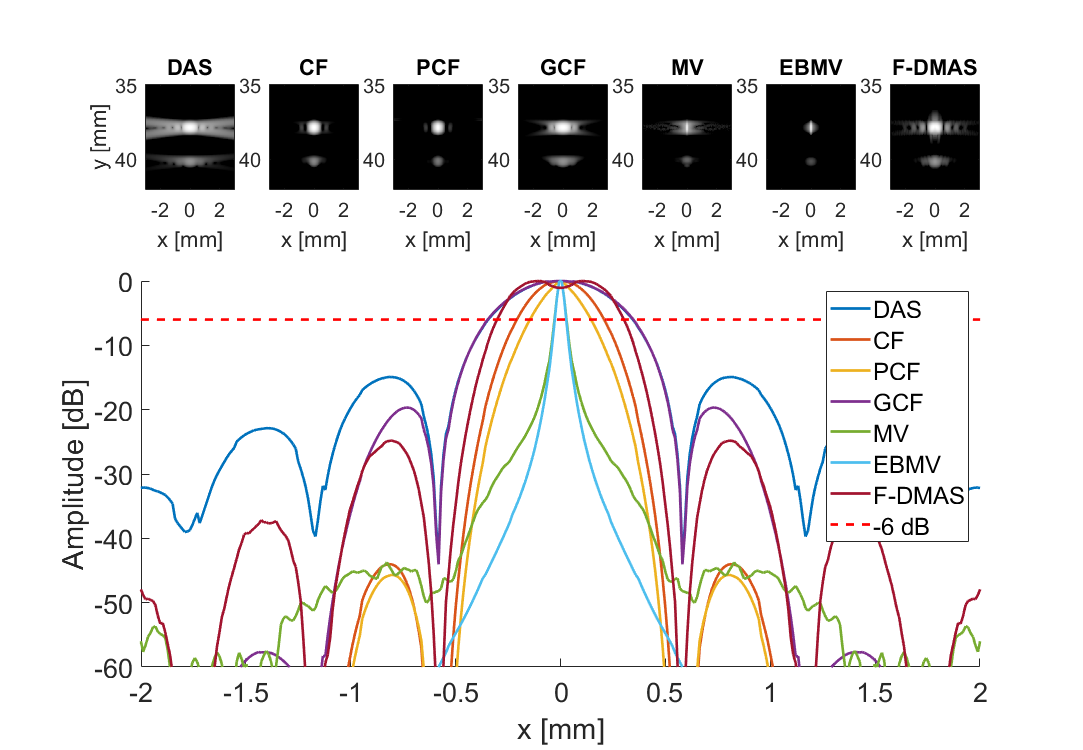
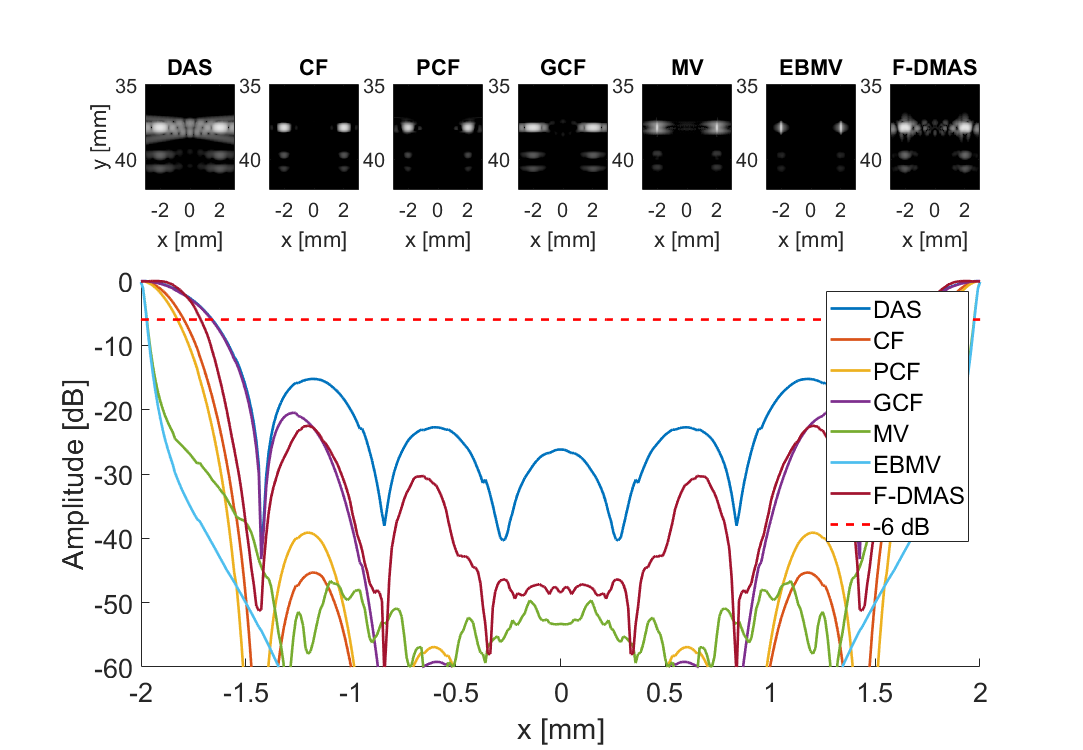
Create plot and movie of images with displacement indicated
frames = [1 2 3 4 5 6 7];
FileName = 'movie_res.mp4';
vidObj = VideoWriter([FileName],'MPEG-4');
vidObj.Quality = 100;
vidObj.FrameRate = 1;
open(vidObj);
idx = 1;
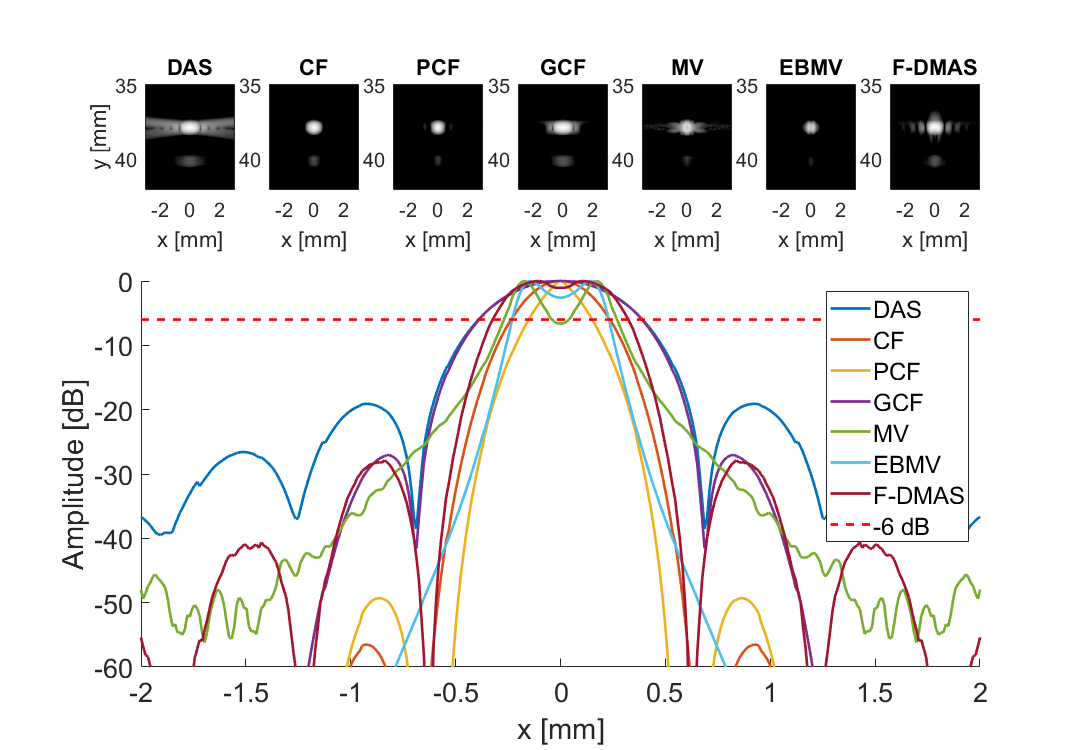
for i = frames
f = figure(100+i);clf;
for m = 1:7
subplot(3,7,m)
imagesc(sca.x_axis*1000,sca.z_axis*1000,img{m}(:,:,i))
ax(m) = gca;
axis image;colormap gray;caxis([-60 0])
title(tags{m});
xlabel('x [mm]');
if(m == 1)
ylabel('y [mm]');
end
set(gca,'FontSize',15)
subplot(3,7,[8:21]); hold all;
plot(sca.x_axis*1000,img{m}(105,:,i)-max(img{m}(105,:,i)),'DisplayName',tags{m},'LineWidth',2,'DisplayName',tags{m})
xlim([-2 2])
ylim([-60 0])
xlabel('x [mm]');
ylabel('Amplitude [dB]');
end
subplot(3,7,[8:21]); hold all;
plot(sca.x_axis*1000,ones(1,length(sca.x_axis))*-6,'r--','DisplayName',tags{m},'LineWidth',2,'DisplayName','-6 dB')
linkaxes(ax);
legend show
set(gcf,'Position',[46 50 1083 750]);
set(gca,'FontSize',20)
if i == 1
mkdir([ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures'])
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'FWHM_PSF_v2'],'eps2c')
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'FWHM_PSF_v2'],'png')
else
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'FWHM_PSF_',...
strrep(num2str(separating_distance(idx)),'.','_'),'_v2'],'eps2c')
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'FWHM_PSF_',...
strrep(num2str(separating_distance(idx)),'.','_'),'_v2'],'png')
idx = idx+1;
drawnow();
writeVideo(vidObj, getframe(f));
end
end
close(vidObj)
Warning: Directory already exists.
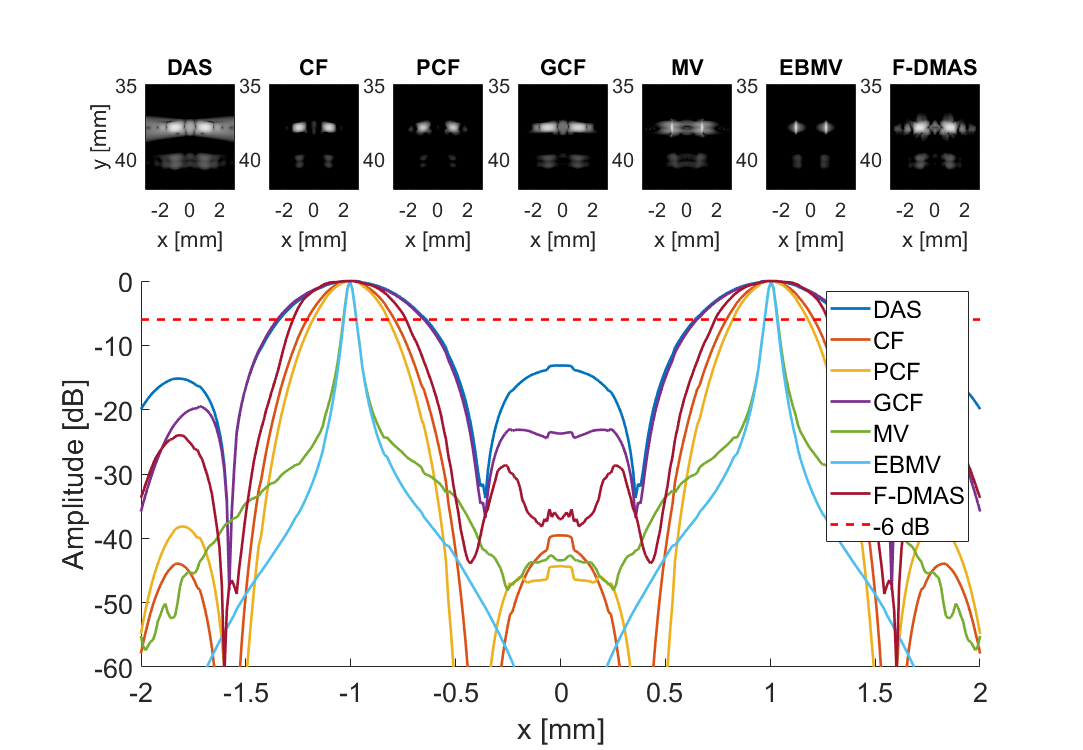
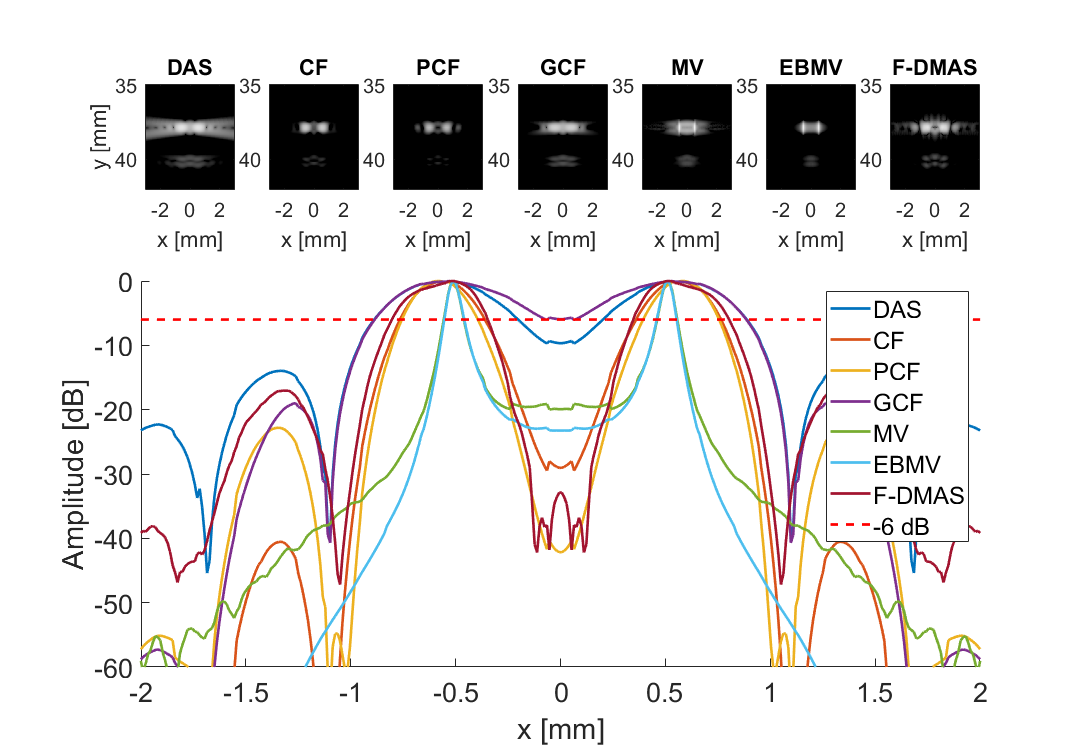
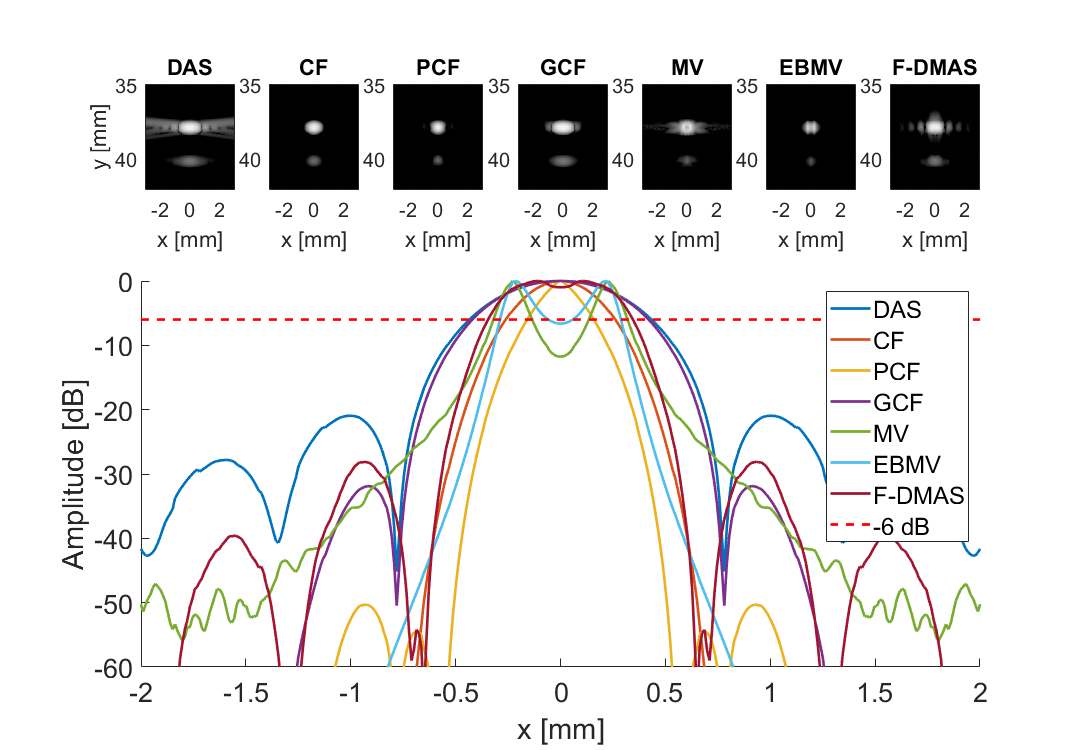
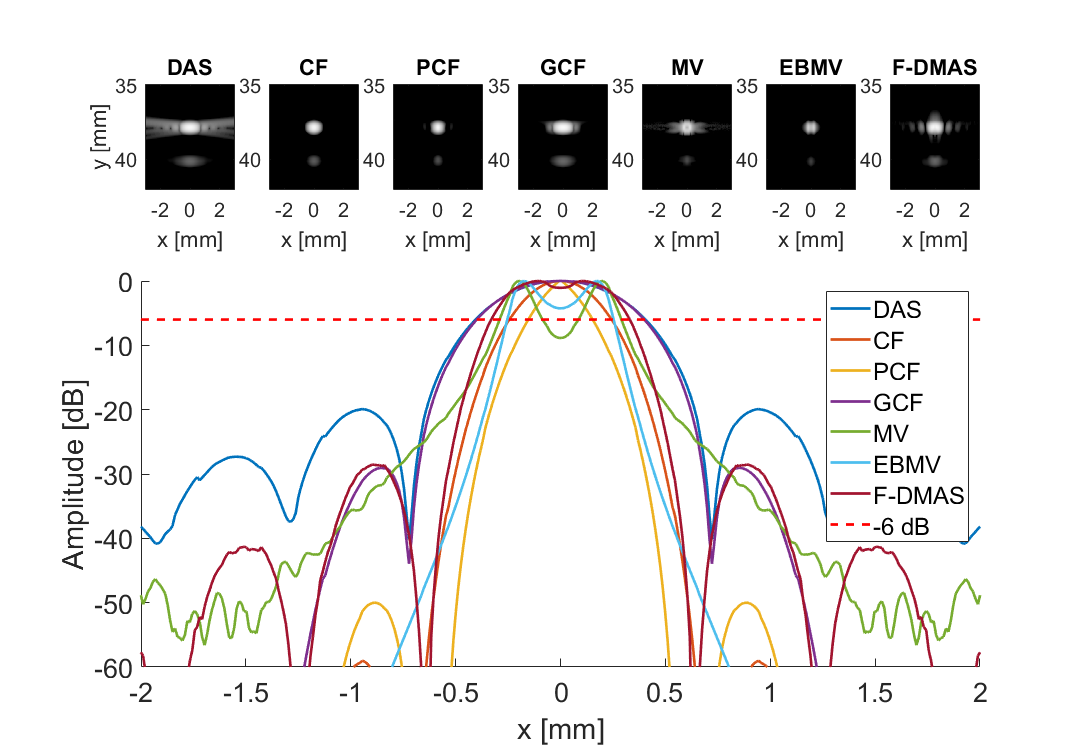
Measure separability
clear separability;
frames = [2:7];
for i = frames
for m= 1:7
normalized_lateral{m} = img{m}(105,:,i)-max(img{m}(105,:,i));
[dummy,idx_start] = min(abs(sca.x_axis*1000-(-2.5)));
[dummy,idx_stop] = min(abs(sca.x_axis*1000-(1)));
[dummy,idx_zero] = min(abs(sca.x_axis*1000));
min_value = normalized_lateral{m}(idx_zero);
[max_value,idx_max] = max(normalized_lateral{m}(idx_start:idx_stop));
separability(m,i-1) = max_value-min_value;
end
end
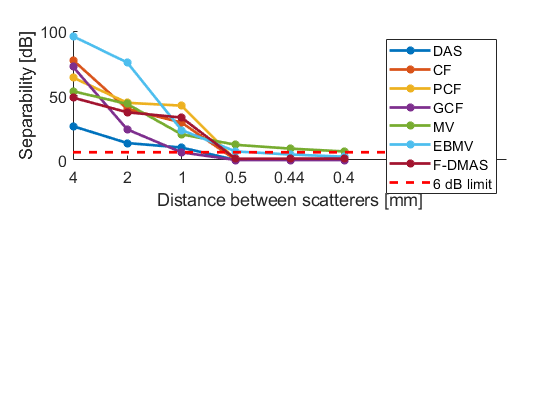
Create summary plots of separability
separability_lim = 6;
f = figure;
subplot(211);hold all;
plot(separability(1,:),'-*','LineWidth',2,'DisplayName',tags{1})
plot(separability(2,:),'-*','LineWidth',2,'DisplayName',tags{2})
plot(separability(3,:),'-*','LineWidth',2,'DisplayName',tags{3})
plot(separability(4,:),'-*','LineWidth',2,'DisplayName',tags{4})
plot(separability(5,:),'-*','LineWidth',2,'DisplayName',tags{5})
plot(separability(6,:),'-*','LineWidth',2,'DisplayName',tags{6})
plot(separability(7,:),'-*','LineWidth',2,'DisplayName',tags{7})
plot([1:7],ones(7,1)*separability_lim,'LineWidth',2,'Color','r',...
'LineStyle','--','DisplayName','6 dB limit')
legend show
xlim([1 9])
xticks([1:6])
xticklabels({num2str(separating_distance(1)) num2str(separating_distance(2))...
num2str(separating_distance(3)) num2str(separating_distance(4)) ...
num2str(separating_distance(5)) num2str(separating_distance(6))})
xlabel('Distance between scatterers [mm]')
ylabel('Separability [dB]');
set(gca,'FontSize',12)
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'separability_1'],'eps2c')
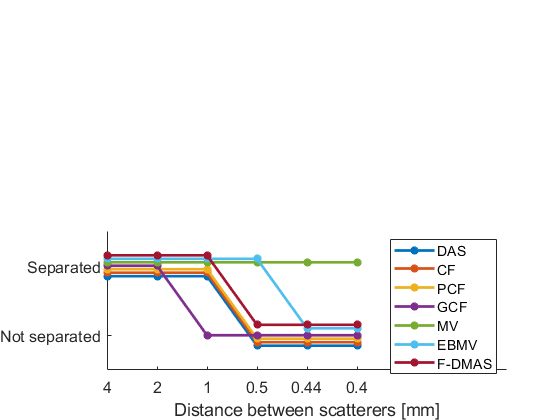
f = figure;
subplot(212);hold all;
plot((separability(1,:)>separability_lim)-0.15,'-*','LineWidth',2,'DisplayName',tags{1})
plot((separability(2,:)>separability_lim)-0.1,'-*','LineWidth',2,'DisplayName',tags{2})
plot((separability(3,:)>separability_lim)-0.05,'-*','LineWidth',2,'DisplayName',tags{3})
plot((separability(4,:)>separability_lim),'-*','LineWidth',2,'DisplayName',tags{4})
plot((separability(5,:)>separability_lim)+0.05,'-*','LineWidth',2,'DisplayName',tags{5})
plot((separability(6,:)>separability_lim)+0.1,'-*','LineWidth',2,'DisplayName',tags{6})
plot((separability(7,:)>separability_lim)+0.15,'-*','LineWidth',2,'DisplayName',tags{7})
ylim([-0.5 1.5])
xticks([1:6])
xticklabels({num2str(separating_distance(1)) num2str(separating_distance(2)) ...
num2str(separating_distance(3)) num2str(separating_distance(4)) num2str(separating_distance(5)) num2str(separating_distance(6))})
xlabel('Distance between scatterers [mm]')
xlim([1 9])
yticks([0 1])
yticklabels({'Not separated','Separated'})
set(gca,'FontSize',12)
legend('show')
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'separability_2'],'eps2c')
saveas(f,[ustb_path,filesep,'publications',filesep,'IUS2020',filesep,...
'Rindal_et_al_Resolution_Measured_as_Separability_Compared_to_FWHM',...
filesep,'figures',filesep,'separability_2'],'png')