Resolution of Delay Multiply And Sum on FI data from an UFF file
by Ole Marius Hoel Rindal olemarius@olemarius.net 28.05.2017
Contents
Setting up file path
To read data from a UFF file the first we need is, you guessed it, a UFF file. We check if it is on the current path and download it from the USTB websever.
clear all; close all; % data location url='http://ustb.no/datasets/'; % if not found downloaded from here local_path = [ustb_path(),'/data/']; % location of example data addpath(local_path); % Choose dataset filename='Alpinion_L3-8_FI_hyperechoic_scatterers.uff'; % check if the file is available in the local path or downloads otherwise tools.download(filename, url, local_path);
Reading channel data from UFF file
uff_file=uff(filename)
channel_data=uff_file.read('/channel_data');
uff_file =
uff with properties:
filename: 'Alpinion_L3-8_FI_hyperechoic_scatterers.uff'
version: 'v1.0.1'
mode: 'append'
verbose: 1
UFF: reading channel_data [uff.channel_data]
UFF: reading sequence [uff.wave]
Processed 256/256
%Print info about the dataset
channel_data.print_info
Name: FI dataset of hyperechoic cyst and points scatterers recorded on an Alpinion scanner with a L3-8 Probe from a CIRS General Purpose Ultrasound Phantom Reference: www.ultrasoundtoolbox.com Author(s): Ole Marius Hoel Rindal <olemarius@olemarius.net> Muyinatu Lediju Bell <mledijubell@jhu.edu> Version: 1.0.2
Define Scan
Define the image coordinates we want to beamform in the scan object. Notice that we need to use quite a lot of samples in the z-direction. This is because the DMAS creates an "artificial" second harmonic signal, so we need high enough sampling frequency in the image to get a second harmonic signal.
z_axis=linspace(25e-3,45e-3,1024).'; sca=uff.linear_scan(); idx = 1; for n=1:numel(channel_data.sequence) sca(n)=uff.linear_scan(channel_data.sequence(n).source.x,z_axis); end
Set up delay part of beamforming
setting up and running the delay part of the beamforming
bmf=beamformer();
bmf.channel_data=channel_data;
bmf.scan=sca;
bmf.receive_apodization.window=uff.window.boxcar;
bmf.receive_apodization.f_number=1.7;
bmf.receive_apodization.apex.distance=Inf;
bmf.transmit_apodization.window=uff.window.none;
b_data=bmf.go({process.delay_matlab_light process.stack});
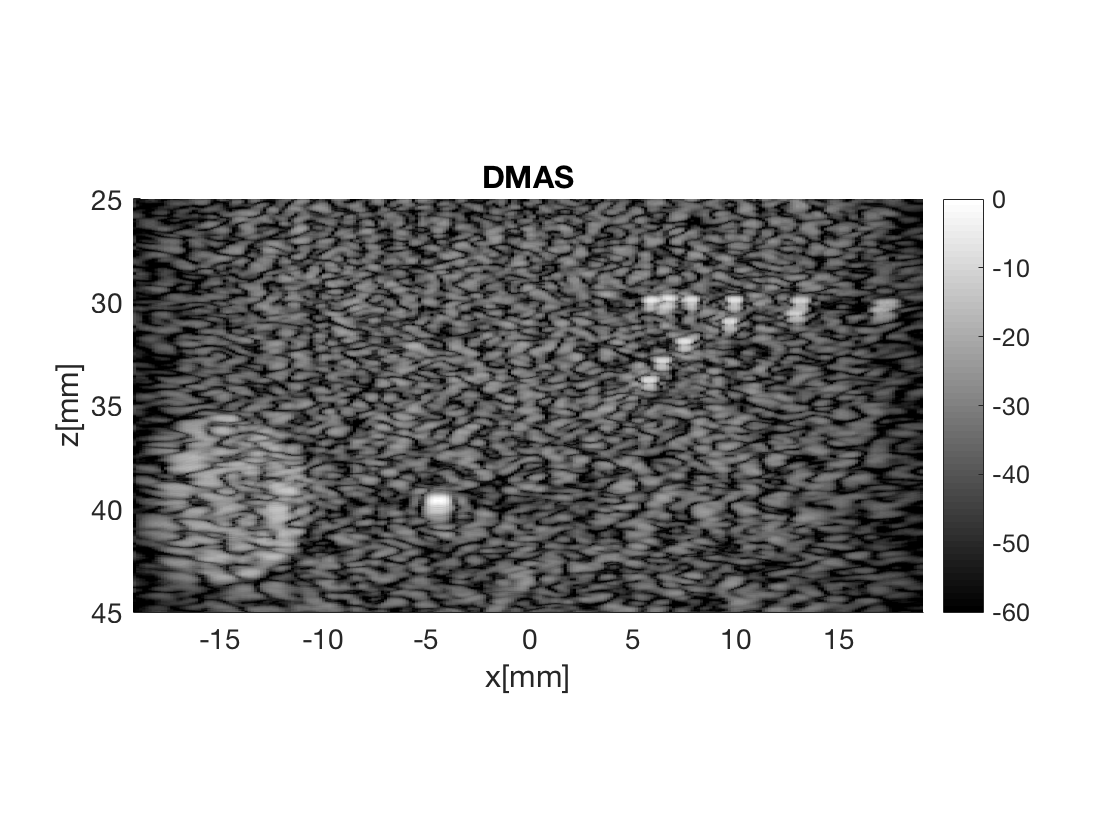
Create the DMAS image using the delay_multiply_and_sum process
dmas = process.delay_multiply_and_sum(); dmas.dimension = dimension.receive(); dmas.receive_apodization = bmf.receive_apodization; dmas.transmit_apodization = bmf.transmit_apodization; dmas.beamformed_data = b_data; dmas.channel_data = channel_data; dmas.scan = sca b_data_dmas = dmas.go(); % Launch beamformer b_data_dmas.plot(100,'DMAS'); % Display image
dmas =
delay_multiply_and_sum with properties:
dimension: receive
channel_data: [1×1 uff.channel_data]
receive_apodization: [1×1 uff.apodization]
transmit_apodization: [1×1 uff.apodization]
scan: [1×256 uff.linear_scan]
beamformed_data: [1×1 uff.beamformed_data]
name: 'Delay Multiply and Sum'
reference: 'Matrone, G., Savoia, A. S., & Magenes, G. (2015...'
implemented_by: {'Ole Marius Hoel Rindal <olemarius@olemarius.net>'}
version: 'v1.0.2'
Warning: If the result looks funky, you might need to tune the filter paramters
of DMAS. Use the plot to check that everything is OK.
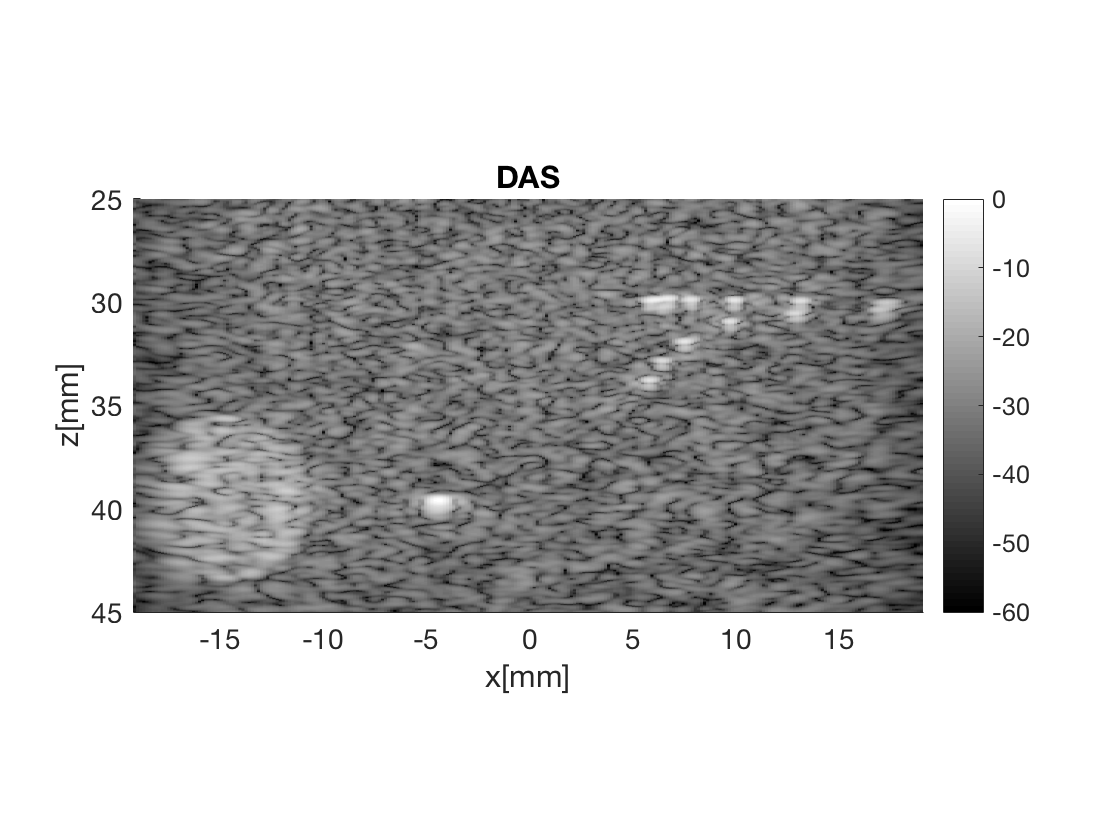
Beamform DAS image
Notice that we just need to sum the data since it is allready delayed
das = process.coherent_compounding();
das.beamformed_data = b_data;
b_data_das=das.go();
b_data_das.plot(2,'DAS');
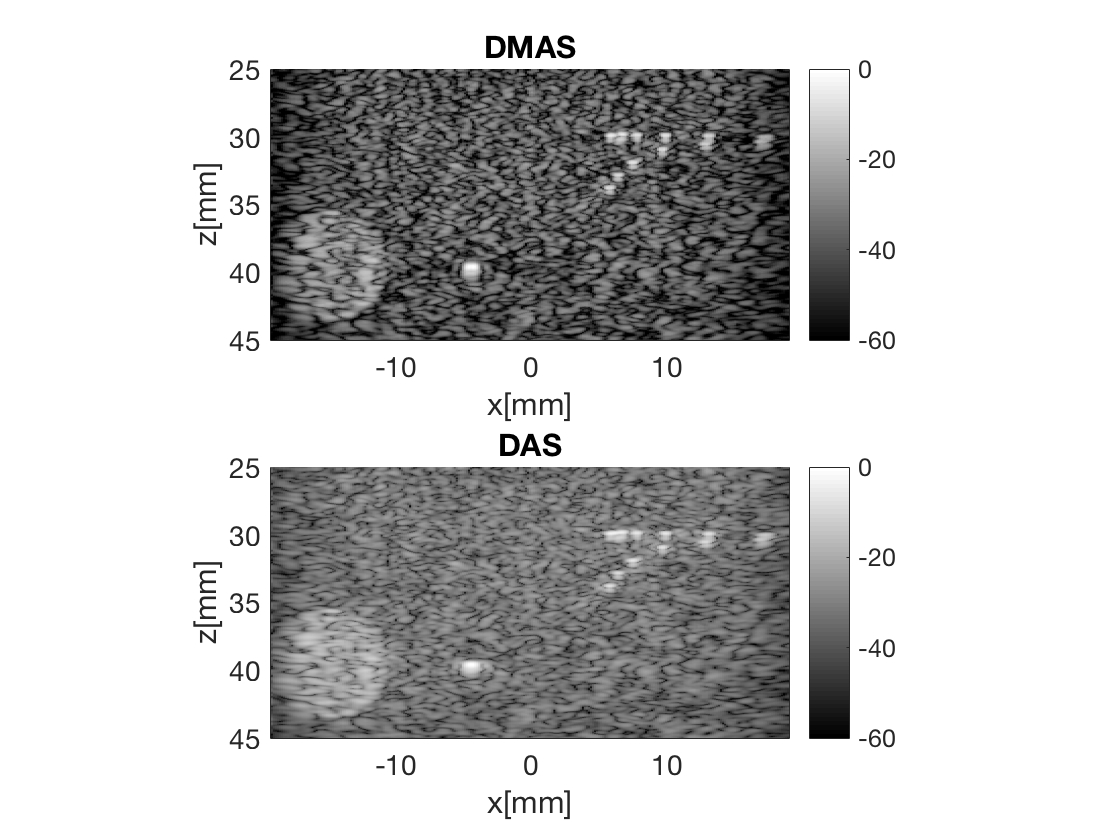
Plot both images in same plot
Plot both in same plot with connected axes, try to zoom!
f3 = figure(3);clf b_data_dmas.plot(subplot(2,1,1),'DMAS'); % Display image ax(1) = gca; b_data_das.plot(subplot(2,1,2),'DAS'); % Display image ax(2) = gca; linkaxes(ax);
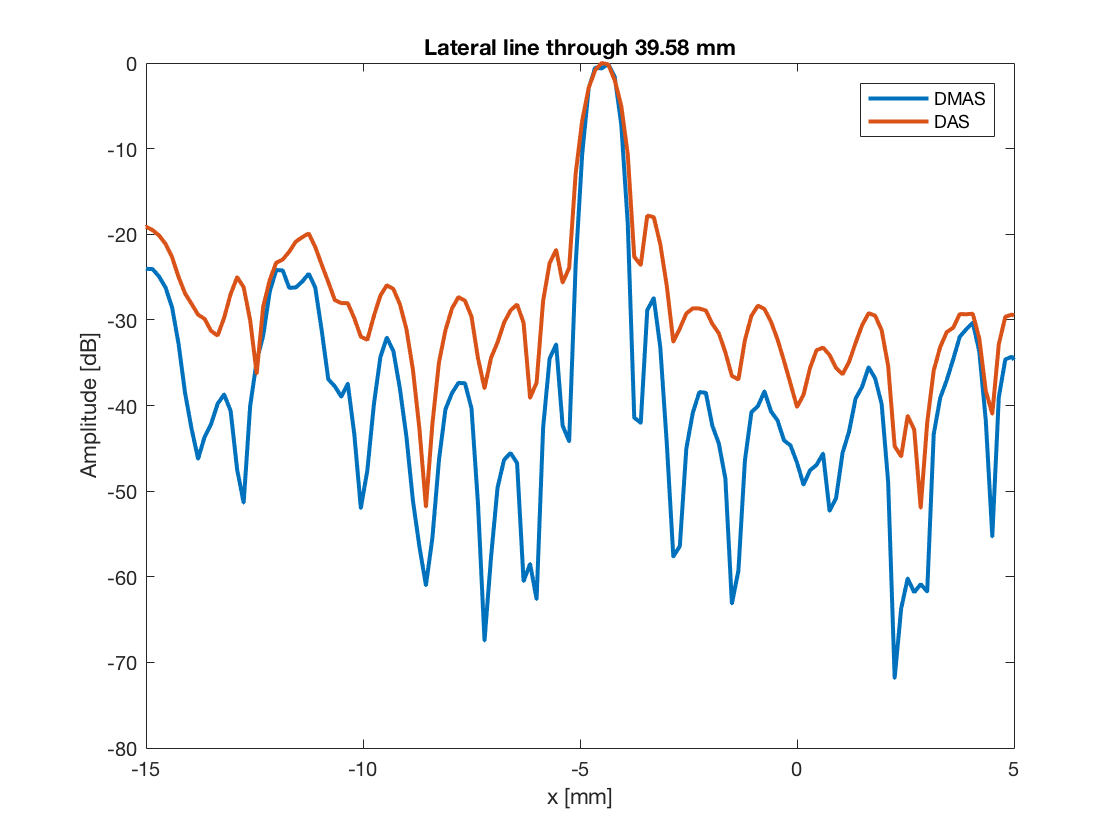
Compare resolution
Plot the lateral line through some of the scatterers
% Let's get the images as a N_z_axis x N_x_axis image
dmas_img = b_data_dmas.get_image();
das_img = b_data_das.get_image();
So that we can plot the line through the group of scatterers
line_idx = 250; figure(4);clf; plot(b_data_dmas.scan.x_axis*10^3,dmas_img(line_idx,:),... 'DisplayName','DMAS','LineWidth',2);hold on; plot(b_data_das.scan.x_axis*10^3,das_img(line_idx,:),... 'DisplayName','DAS','LineWidth',2); xlabel('x [mm]');xlim([0 20]);ylabel('Amplitude [dB]');legend show title(sprintf('Lateral line through %.2f mm',... b_data_dmas.scan.z_axis(line_idx)*10^3));
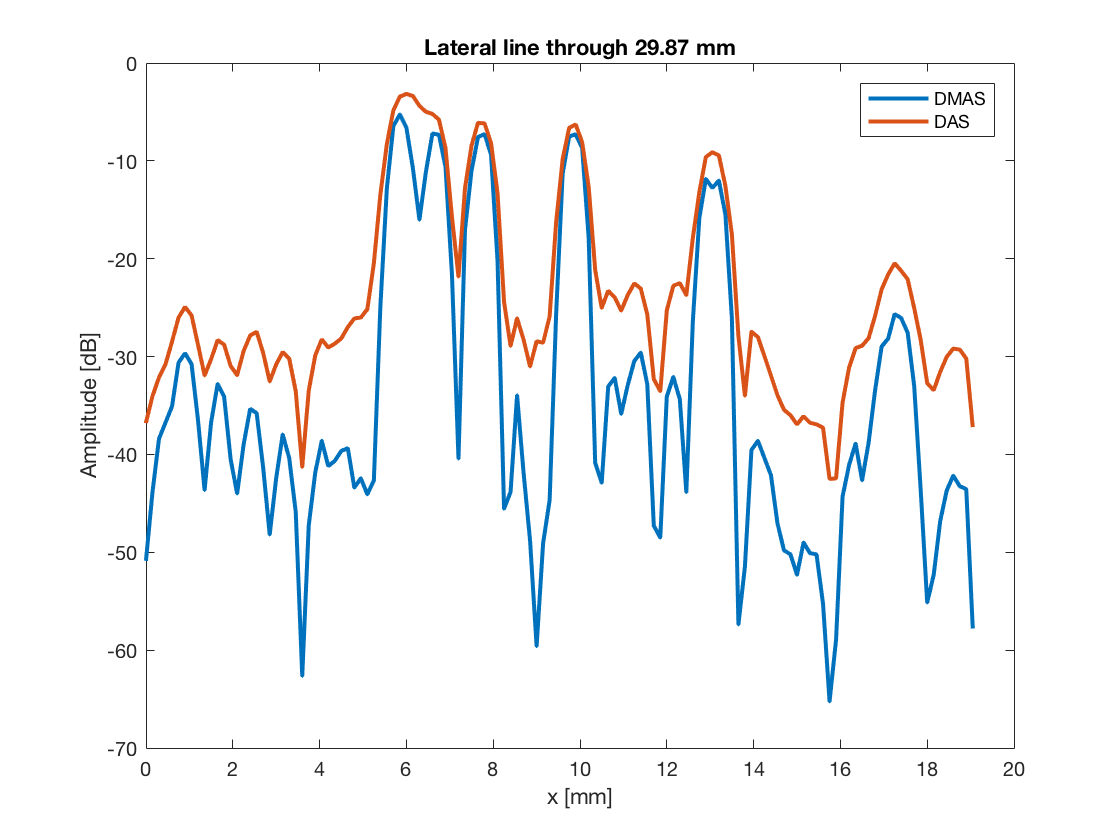
%So that we can plot the line through the aingle scatterer line_idx = 747; figure(5);clf; plot(b_data_dmas.scan.x_axis*10^3,dmas_img(line_idx,:),... 'DisplayName','DMAS','LineWidth',2);hold on; plot(b_data_das.scan.x_axis*10^3,das_img(line_idx,:),... 'DisplayName','DAS','LineWidth',2); xlabel('x [mm]');xlim([-15 5]);ylabel('Amplitude [dB]');legend show title(sprintf('Lateral line through %.2f mm',... b_data_dmas.scan.z_axis(line_idx)*10^3));