Create the figures from the simulated dynamic range phantom
For the publication Rindal, O. M. H., Austeng, A., Fatemi, A., & Rodriguez-Molares, A. (2019). The effect of dynamic range alterations in the estimation of contrast. Submitted to IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control.
NB! You need to run the create_figures_from_experimental.m first since we are using the stored experimental gradient values and contrast values to plot both the simulated and the experimental in the same plot.
Author: Ole Marius Hoel Rindal olemarius@olemarius.net 05.06.18 updated for revised version of manuscript 07.03.19
Contents
- Load the data from the uff file
- Print authorship and citation details for the dataset
- DAS - Delay-And-Sum
- GLT - Gray Level Transform
- CF - Coherence Factor
- PCF - Phase Coherence Factor
- GCF - Generalized Coherence Factor
- MV - Capon's Minimum Variance
- F-DMAS - Filtered-Delay-Multiply-and-Sum
- EBMV - Eigenspace Based Minimum Variance
- Putting the dB images and the signal in a cell array
- Plot the lateral and axial gradient together with the experimental lateral gradient
- Plot the lateral line through the boxes
- Calibrate images
- Plot the lateral line through the boxes after calibration
- Measure contrast and plot together with the experimental
- Run Dynamic Range Test
Load the data from the uff file
clear all; close all; addpath([ustb_path,filesep,'publications',filesep,'DynamicRange',filesep,'functions',filesep]) filename = 'FieldII_STAI_dynamic_range.uff'; url='http://ustb.no/datasets/'; % if not found downloaded from here % checks if the data is in your data path, and downloads it otherwise. % The defaults data path is under USTB's folder, but you can change this % by setting an environment variable with setenv(DATA_PATH,'the_path_you_want_to_use'); tools.download(filename, url, data_path); channel_data = uff.channel_data(); channel_data.read([data_path,filesep,filename],'/channel_data'); b_data_das = uff.beamformed_data(); b_data_cf = uff.beamformed_data(); b_data_pcf = uff.beamformed_data(); b_data_gcf = uff.beamformed_data(); b_data_mv = uff.beamformed_data(); b_data_ebmv = uff.beamformed_data(); b_data_dmas = uff.beamformed_data(); b_data_weights = uff.beamformed_data(); b_data_das.read([data_path,filesep,filename],'/b_data_das'); b_data_cf.read([data_path,filesep,filename],'/b_data_cf'); b_data_pcf.read([data_path,filesep,filename],'/b_data_pcf'); b_data_gcf.read([data_path,filesep,filename],'/b_data_gcf'); b_data_mv.read([data_path,filesep,filename],'/b_data_mv'); b_data_ebmv.read([data_path,filesep,filename],'/b_data_ebmv'); b_data_dmas.read([data_path,filesep,filename],'/b_data_dmas'); b_data_weights.read([data_path,filesep,filename],'/b_data_weights'); weights = b_data_weights.get_image('none'); mkdir([ustb_path,filesep,'publications/DynamicRange/figures/simulation/']); mkdir([ustb_path,filesep,'publications/DynamicRange/figures/simulation/gradient/']); mkdir([ustb_path,filesep,'publications/DynamicRange/figures/simulation/calibrated/']); mkdir([ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/']);
UFF: reading channel_data [uff.channel_data] UFF: reading sequence [uff.wave] [====================] 100% UFF: reading /b_data_das [uff.beamformed_data] UFF: reading /b_data_cf [uff.beamformed_data] UFF: reading /b_data_pcf [uff.beamformed_data] UFF: reading /b_data_gcf [uff.beamformed_data] UFF: reading /b_data_mv [uff.beamformed_data] UFF: reading /b_data_ebmv [uff.beamformed_data] UFF: reading /b_data_dmas [uff.beamformed_data] UFF: reading /b_data_weights [uff.beamformed_data] Warning: Directory already exists. Warning: Directory already exists. Warning: Directory already exists. Warning: Directory already exists.
Print authorship and citation details for the dataset
channel_data.print_authorship
Name: v5 Simulated dynamic range phantom. Created with Field II. See the reference for details Reference: Rindal, O. M. H., Austeng, A., Fatemi, A., & Rodriguez-Molares, A. (2018). The effect of dynamic range transformations in the estimation of contrast. Submitted to IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. Author(s): Ole Marius Hoel Rindal <omrindal@ifi.uio.no Alfonso Rodriguez-Molares <alfonso.r.molares@ntnu.no> Version: 1.0.1
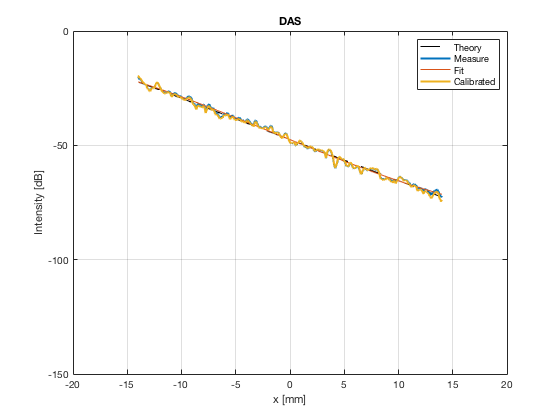
DAS - Delay-And-Sum
Create the individual images and the zoomed images on the boxes
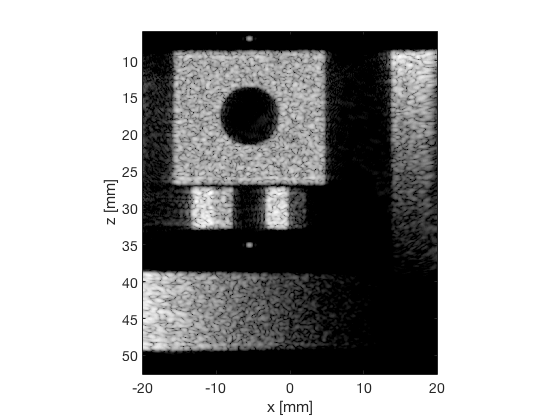
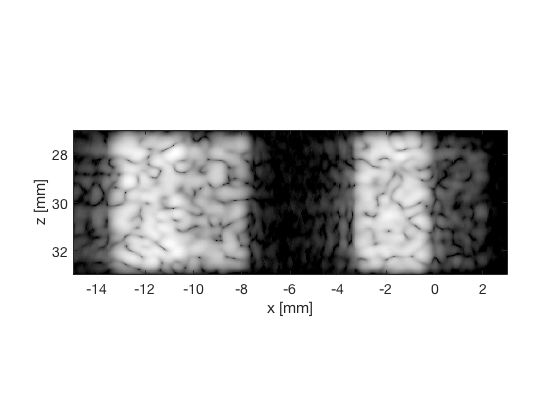
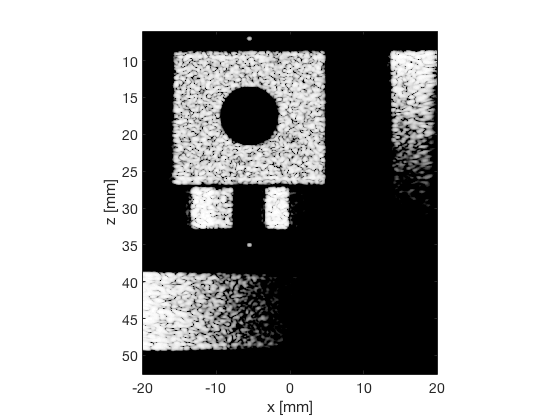
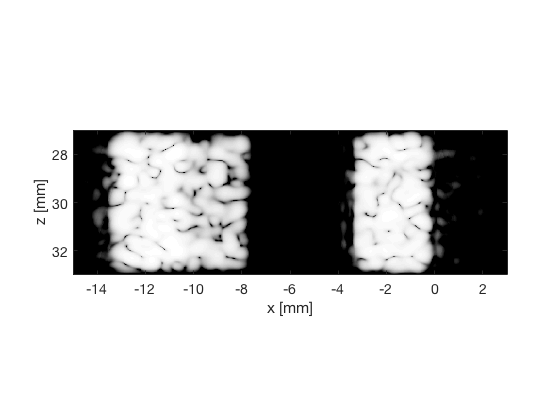
das_img = b_data_das.get_image('none').*weights; % Compensation weighting b_data_das.data = das_img(:); %Putting the weighted data back... das_img = db(abs(das_img./max(das_img(:)))); % Normalize on max f1 = figure(1);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,das_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14); saveas(f1,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/DAS'],'eps2c') saveas(f1,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/DAS'],'png') f2 = figure(2);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,das_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f2,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/DAS_zoomed'],'eps2c')
GLT - Gray Level Transform
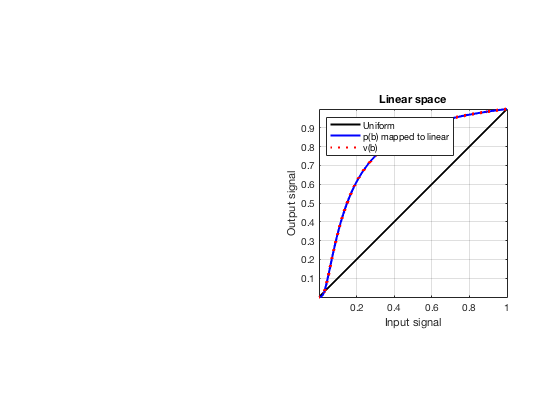
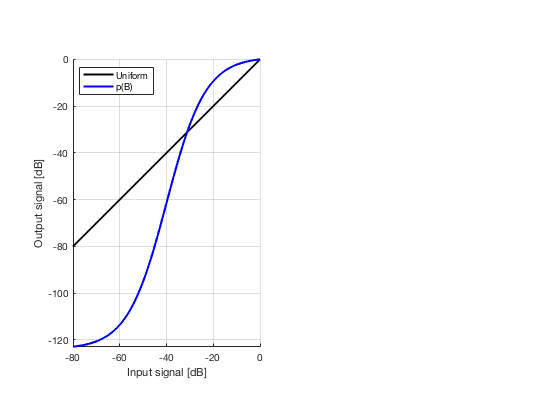
glt_s = postprocess.scurve_gray_level_transform(); glt_s.a = 0.12; glt_s.b = -40; glt_s.c = 0.008; glt_s.plot_functions = 1; glt_s.input = b_data_das; glt_s.scan = b_data_das.scan; b_data_glt = glt_s.go(); glt_img = b_data_glt.get_image('none'); % Compensation weighting b_data_glt.data = glt_img(:); glt_img = db(abs(glt_img./max(glt_img(:)))+eps); % Normalize on max f100 = figure(100);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,glt_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) saveas(f100,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/GLT'],'eps2c') f101 = figure(101);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,glt_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f101,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/GLT_zoomed'],'eps2c')
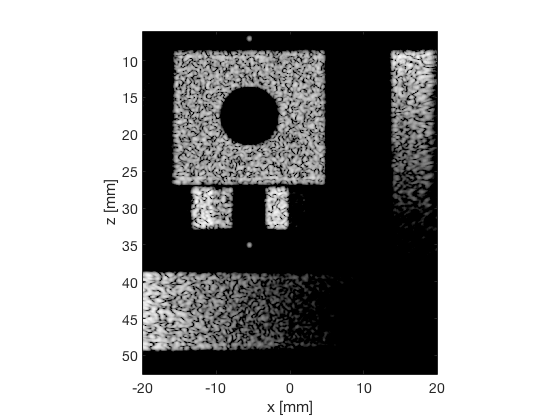
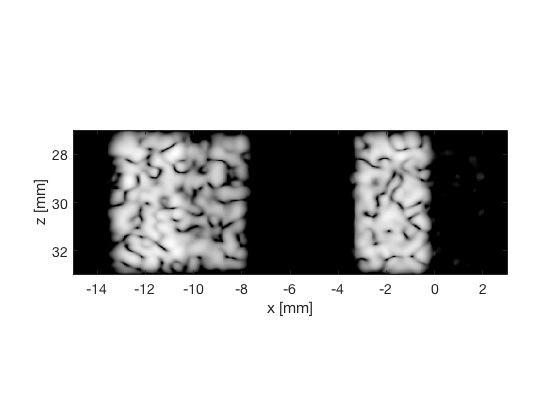
CF - Coherence Factor
cf_img = b_data_cf.get_image('none').*weights; b_data_cf.data = cf_img(:); cf_img = db(abs(cf_img./max(cf_img(:)))); % Normalize on max f2 = figure(2); imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,cf_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) saveas(f2,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/CF'],'eps2c') f22 = figure(22); imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,cf_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f22,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/CF_zoomed'],'eps2c')
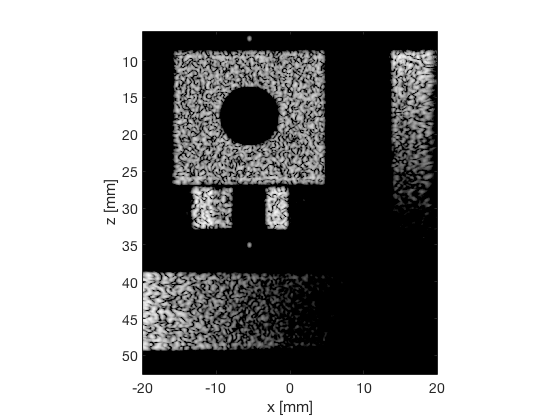
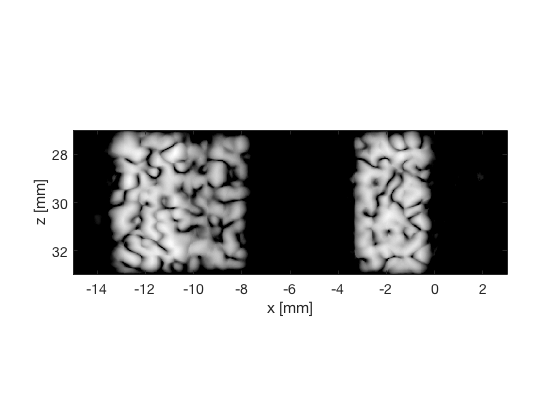
PCF - Phase Coherence Factor
pcf_img = b_data_pcf.get_image('none').*weights; b_data_pcf.data = pcf_img(:); b_data_pcf.data(isinf(b_data_pcf.data)) = eps; b_data_pcf.data(b_data_pcf.data==0) = eps; pcf_img(isinf(pcf_img)) = eps; pcf_img(pcf_img==0) = eps; pcf_img = db(abs(pcf_img./max(pcf_img(:)))+eps); f3 = figure(3);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,pcf_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) saveas(f3,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/PCF'],'eps2c') f33 = figure(33);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,pcf_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f33,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/PCF_zoomed'],'eps2c')
GCF - Generalized Coherence Factor
gcf_img = b_data_gcf.get_image('none').*weights; b_data_gcf.data = gcf_img(:); gcf_img = db(abs(gcf_img./max(gcf_img(:)))); f4 = figure(4);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,gcf_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) saveas(f4,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/GCF'],'eps2c') f44 = figure(44); imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,gcf_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f44,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/GCF_zoomed'],'eps2c')
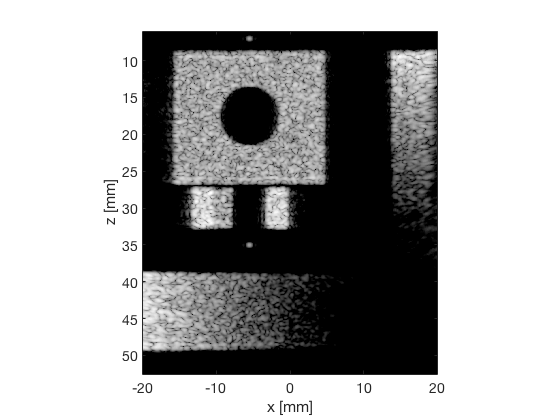
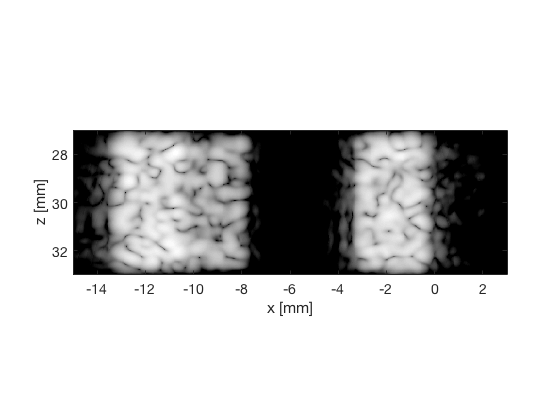
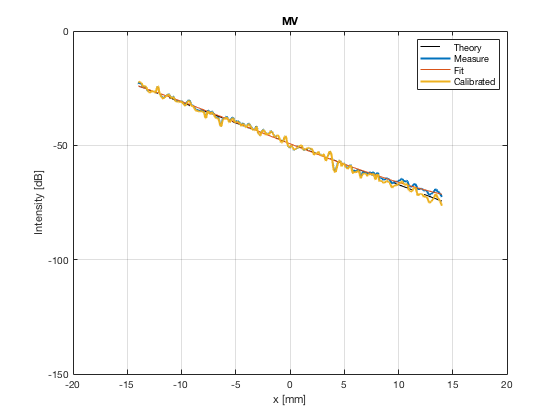
MV - Capon's Minimum Variance
mv_img = b_data_mv.get_image('none').*weights; b_data_mv.data = mv_img(:); mv_img = db(abs(mv_img./max(mv_img(:)))); f6 = figure(6);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,mv_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) saveas(f6,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/MV'],'eps2c') f66 = figure(66);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,mv_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f66,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/MV_zoomed'],'eps2c')
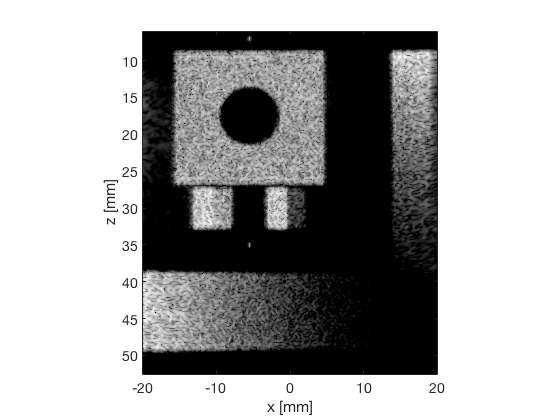
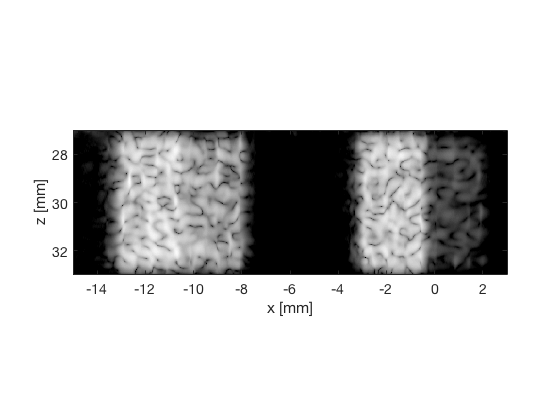
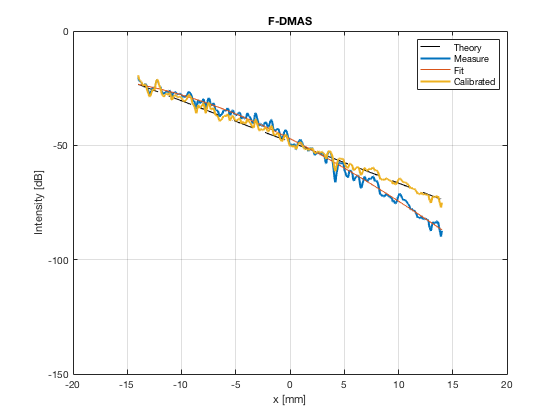
F-DMAS - Filtered-Delay-Multiply-and-Sum
dmas_img = b_data_dmas.get_image('none').*weights; b_data_dmas.data = dmas_img(:); dmas_img = db(abs(dmas_img./max(dmas_img(:)))); f7 = figure(7);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,dmas_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) saveas(f7,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/DMAS'],'eps2c') f77 = figure(77);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,dmas_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f77,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/DMAS_zoomed'],'eps2c')
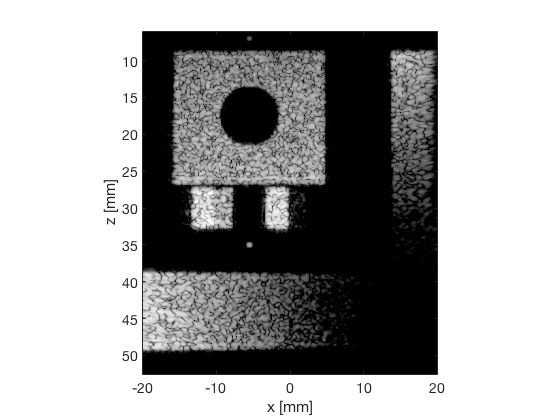
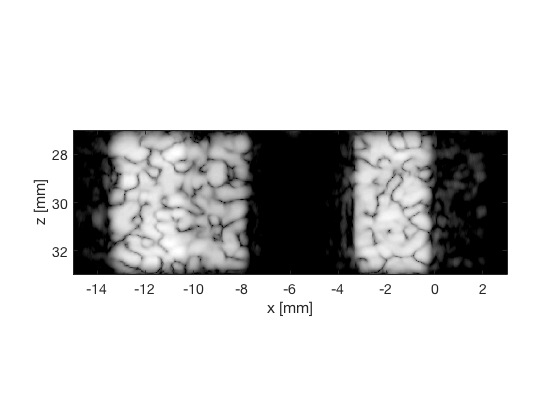
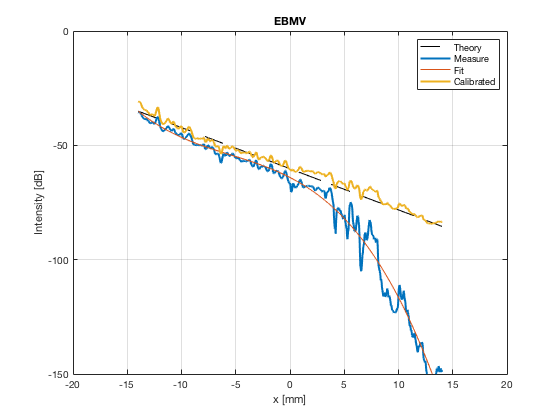
EBMV - Eigenspace Based Minimum Variance
ebmv_img = b_data_ebmv.get_image('none').*weights; b_data_ebmv.data = ebmv_img(:); ebmv_img = db(abs(ebmv_img./max(ebmv_img(:)))); f8 = figure(8);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,ebmv_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) saveas(f8,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/EBMV'],'eps2c') f88 = figure(88);clf; imagesc(b_data_das.scan.x_axis*1000,b_data_das.scan.z_axis*1000,ebmv_img); colormap gray;caxis([-60 0]);axis image;xlabel('x [mm]');ylabel('z [mm]'); set(gca,'FontSize',14) axis([-15 3 27 33]); saveas(f88,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/EBMV_zoomed'],'eps2c')
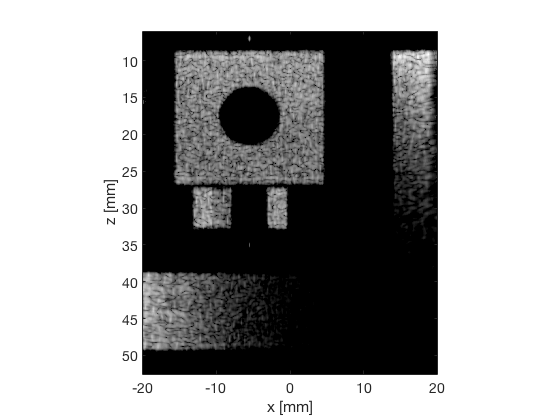
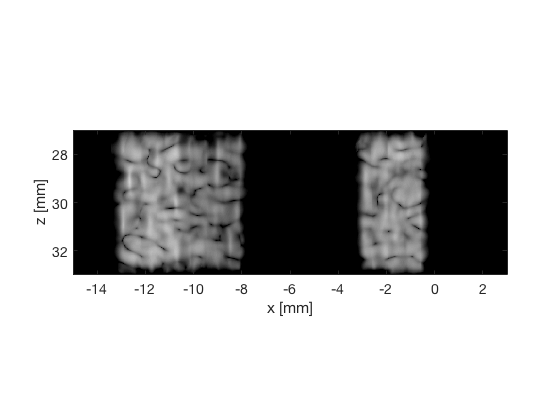
Putting the dB images and the signal in a cell array
addpath([ustb_path,'/publications/DynamicRange/functions']); image.all{1} = das_img; image.all_signal{1} = double(b_data_das.get_image('none')); image.all_signal{1} = (image.all_signal{1}./max(image.all_signal{1}(:))); image.tags{1} = 'DAS'; image.all{2} = mv_img; image.all_signal{2} = double(b_data_mv.get_image('none')); image.all_signal{2} = (image.all_signal{2}./max(image.all_signal{2}(:))); image.tags{2} = 'MV'; image.all{3} = ebmv_img; image.all_signal{3} = double(b_data_ebmv.get_image('none')); image.all_signal{3} = (image.all_signal{3}./max(image.all_signal{3}(:))); image.tags{3} = 'EBMV'; image.all{4} = dmas_img; image.all_signal{4} = double(b_data_dmas.get_image('none')); image.all_signal{4} = (image.all_signal{4}./max(image.all_signal{4}(:))); image.tags{4} = 'F-DMAS'; image.all{5} = cf_img; image.all_signal{5} = double(b_data_cf.get_image('none')); image.all_signal{5} = (image.all_signal{5}./max(image.all_signal{5}(:))); image.tags{5} = 'CF'; image.all{6} = gcf_img; image.all_signal{6} = double(b_data_gcf.get_image('none')); image.all_signal{6} = (image.all_signal{6}./max(image.all_signal{6}(:))); image.tags{6} = 'GCF'; image.all{7} = pcf_img; image.all_signal{7} = double(b_data_pcf.get_image('none')); image.all_signal{7}(isinf(image.all_signal{7})) = eps; image.all_signal{7} = (image.all_signal{7}./max(image.all_signal{7}(:))); image.tags{7} = 'PCF'; image.all{8} = glt_img; image.all_signal{8} = double(b_data_glt.get_image('none')); image.all_signal{8} = (image.all_signal{8}./max(image.all_signal{8}(:))); image.tags{8} = 'GLT'; % Using "linspecer" colors colors= [0.9047 0.1918 0.1988; ... 0.2941 0.5447 0.7494; ... 0.3718 0.7176 0.3612; ... 1.0000 0.5482 0.1000; ... 0.8650 0.8110 0.4330; ... 0.6859 0.4035 0.2412; ... 0.9718 0.5553 0.7741; ... 0.6400 0.6400 0.6400];
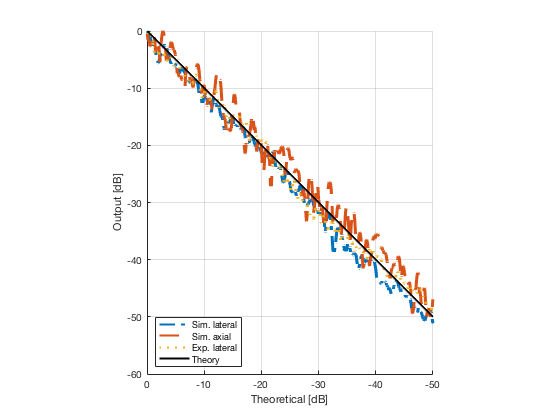
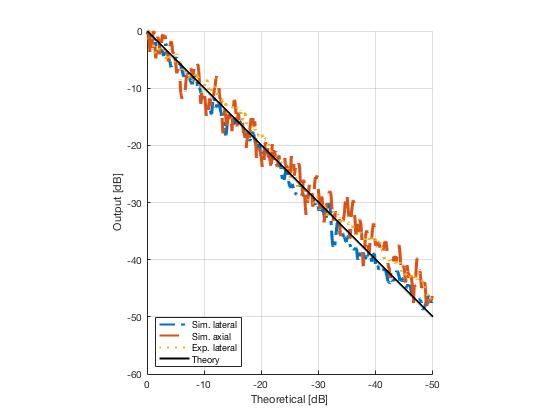
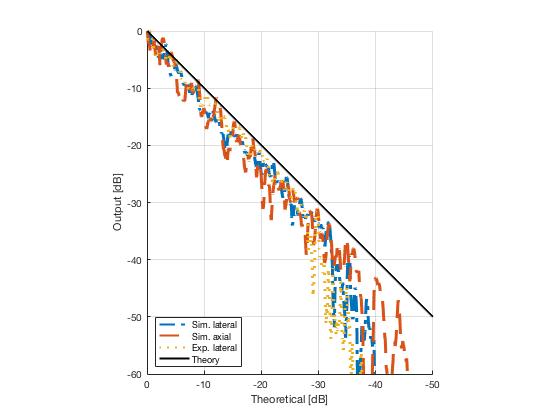
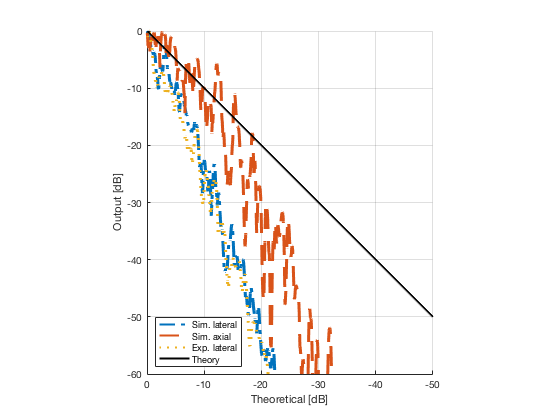
Plot the lateral and axial gradient together with the experimental lateral gradient
image_experimental = load([ustb_path,'/publications/DynamicRange/','Experimental.mat'],'image'); image_experimental = image_experimental.image; b_data_das_exp = load([ustb_path,'/publications/DynamicRange/','Experimental.mat'],'b_data_das'); b_data_das_exp = b_data_das_exp.b_data_das; x_axis_grad_start = 14; [meanLinesLateralExp,~] = getMeanLateralLines(b_data_das_exp,image_experimental,39,48.5,-x_axis_grad_start,x_axis_grad_start); gradient = -1.8; mask_lateral_exp=abs(b_data_das_exp.scan.x_axis)<x_axis_grad_start*10^-3; %theory_lateral_exp=-40*(b_data_das_exp.scan.x_axis(mask_lateral_exp)+x_axis_grad_start*10^-3)/24.1e-3; theory_lateral_exp = (gradient*(b_data_das_exp.scan.x_axis(mask_lateral_exp)+x_axis_grad_start*10^-3))*10^3; [meanLinesLateral,x_axis] = getMeanLateralLines(b_data_das,image,39,48.5,-x_axis_grad_start,x_axis_grad_start); mask_lateral=abs(b_data_das.scan.x_axis)<x_axis_grad_start*10^-3; theory_lateral = (gradient*(b_data_das.scan.x_axis(mask_lateral)+x_axis_grad_start*10^-3))*10^3; [meanLines_axial,z_axis] = getMeanAxialLines(b_data_das,image,10,38,15,18.5); mask_axial= b_data_das.scan.z_axis<38e-3 & b_data_das.scan.z_axis>10e-3; theory_axial = (gradient*(b_data_das.scan.z_axis(mask_axial)-10e-3))*10^3; for i = 1:length(image.all) f88 = figure(8888+i);clf;hold all; plot(theory_lateral,meanLinesLateral.all{i}-max(meanLinesLateral.all{i}),'LineStyle','-.','Linewidth',2,'DisplayName','Sim. lateral'); plot(theory_axial,meanLines_axial.all{i}-max(meanLines_axial.all{i}),'Linestyle','--','Linewidth',2,'DisplayName','Sim. axial'); plot(theory_lateral_exp,meanLinesLateralExp.all{i}-max(meanLinesLateralExp.all{i}),'Linestyle',':','Linewidth',2,'DisplayName','Exp. lateral'); plot(theory_lateral,theory_lateral,'LineStyle','-','Linewidth',2,'Color','k','DisplayName','Theory'); set(gca, 'XDir','reverse');%title(image.tags{i}); axis image legend('show','Location','sw'); ylim([-60 0]) xlim([-50 0]) grid on xlabel('Theoretical [dB]'); ylabel('Output [dB]'); saveas(f88,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/gradient/',image.tags{i}],'eps2c') %saveas(f88,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/gradient/',image.tags{i}],'png') end
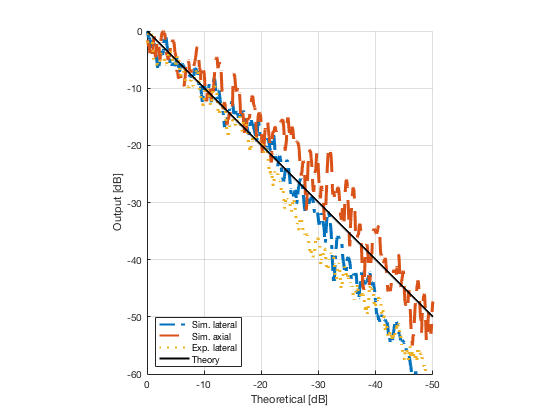
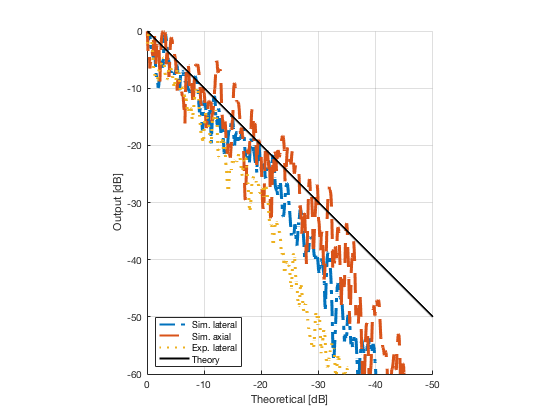
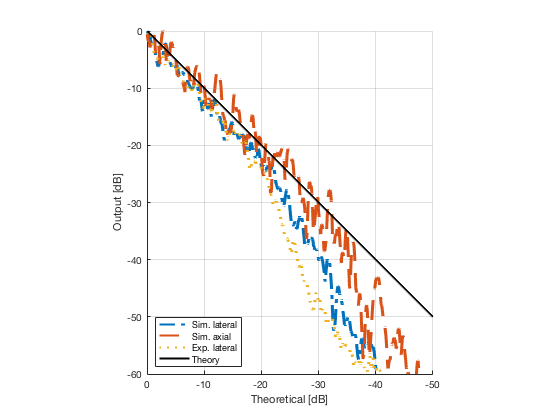
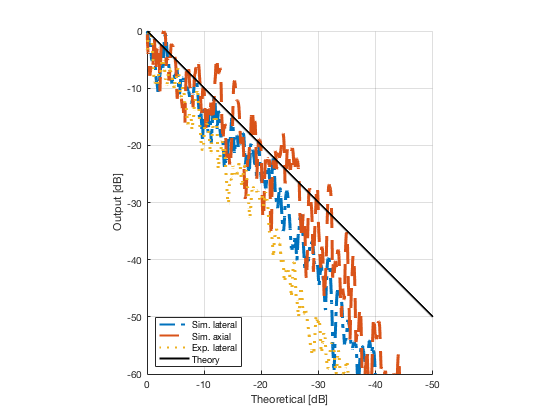
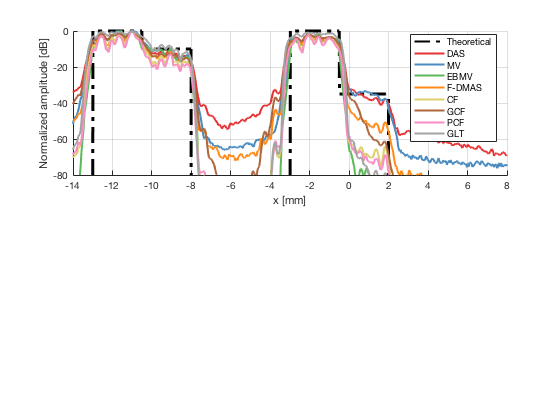
Plot the lateral line through the boxes
[meanLines,x_axis] = getMeanLateralLines(b_data_das,image,27.5,32.5,b_data_das.scan.x(1)*10^3,b_data_das.scan.x(end)*10^3); theoretical = [-100 ones(1,255)*0 ones(1,255)*-10 -100 ones(1,256)*-100 ones(1,256)*-100 -100 ones(1,255)*0 ones(1,255)*-35 -100]; x_axis_theoretical = linspace(-13,2,1536); f89 = figure(89);clf;hold all; subplot(211);hold on; plot(x_axis_theoretical,theoretical,'LineStyle','-.','Linewidth',2,'DisplayName','Theoretical','Color',[0 0 0]); plot(x_axis,meanLines.all{1}-max(meanLines.all{1}),'Linewidth',2,'DisplayName',image.tags{1},'Color',colors(1,:)); plot(x_axis,meanLines.all{2}-max(meanLines.all{2}),'Linewidth',2,'DisplayName',image.tags{2},'Color',colors(2,:)); plot(x_axis,meanLines.all{3}-max(meanLines.all{3}),'Linewidth',2,'DisplayName',image.tags{3},'Color',colors(3,:)); plot(x_axis,meanLines.all{4}-max(meanLines.all{4}),'Linewidth',2,'DisplayName',image.tags{4},'Color',colors(4,:)); plot(x_axis,meanLines.all{5}-max(meanLines.all{5}),'Linewidth',2,'DisplayName',image.tags{5},'Color',colors(5,:)); plot(x_axis,meanLines.all{6}-max(meanLines.all{6}),'Linewidth',2,'DisplayName',image.tags{6},'Color',colors(6,:)); plot(x_axis,meanLines.all{7}-max(meanLines.all{7}),'Linewidth',2,'DisplayName',image.tags{7},'Color',colors(7,:)); plot(x_axis,meanLines.all{8}-max(meanLines.all{8}),'Linewidth',2,'DisplayName',image.tags{8},'Color',colors(8,:)); ylim([-80 0]); xlim([-14 8]); legend('Location','best'); grid on; ylabel('Normalized amplitude [dB]'); xlabel('x [mm]');
saveas(f89,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/boxes'],'eps2c')
Calibrate images
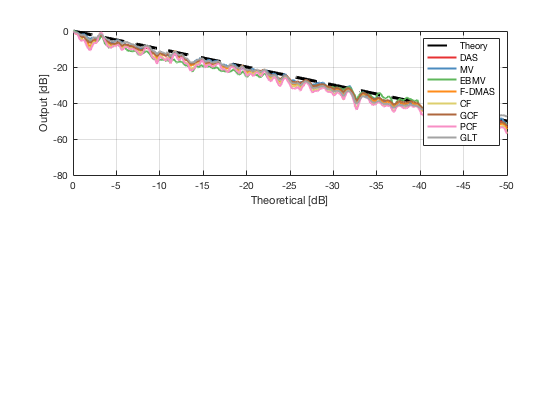
[image_calibrated_on_theoretical] = calibrateImage_on_theoretical(b_data_das,image,40,48.5,-14,14,3,1.8,colors); x_axis_grad_start = 14; mask_lateral=abs(b_data_das.scan.x_axis)<x_axis_grad_start*10^-3; theory_lateral = (-1.8*(b_data_das.scan.x_axis(mask_lateral)+x_axis_grad_start*10^-3))*10^3; [meanLines_calibrated,~] = getMeanLateralLines(b_data_das,image_calibrated_on_theoretical,39,48.5,-x_axis_grad_start,x_axis_grad_start); f10 = figure(77);clf;hold all; subplot(211); plot(theory_lateral,theory_lateral,'k--','DisplayName','Theory','linewidth',2); hold on; grid on; for i = 1:length(image_calibrated_on_theoretical.all) plot(theory_lateral,meanLines_calibrated.all{i}-max(meanLines_calibrated.all{i}),'linewidth',2,'DisplayName',image.tags{i},'Color',colors(i,:)); end xlabel('Theoretical [dB]'); ylabel('Output [dB]'); set(gca, 'XDir','reverse'); xlim([-50 0]) ylim([-80 0]) legend show saveas(f10,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/calibrated/calibrated_all_curves_on_theoretical'],'eps2c')
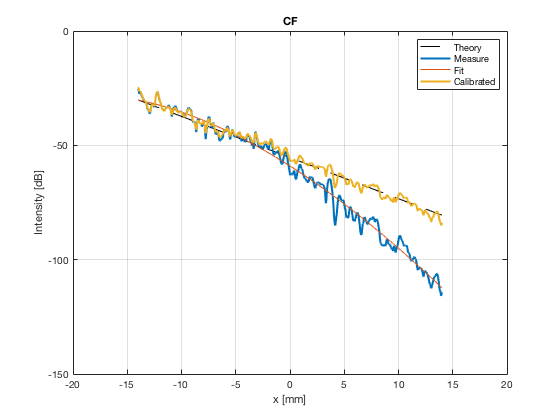
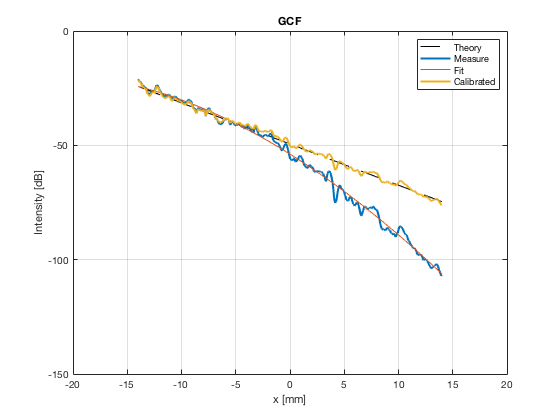
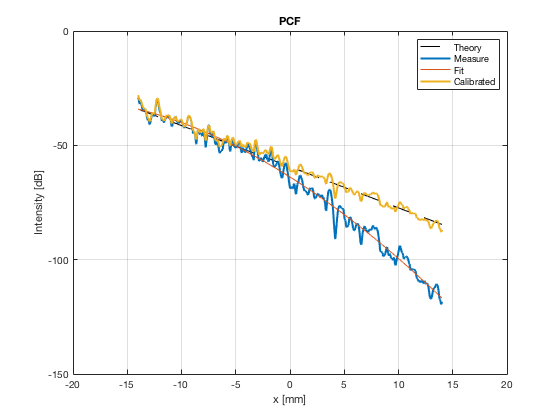
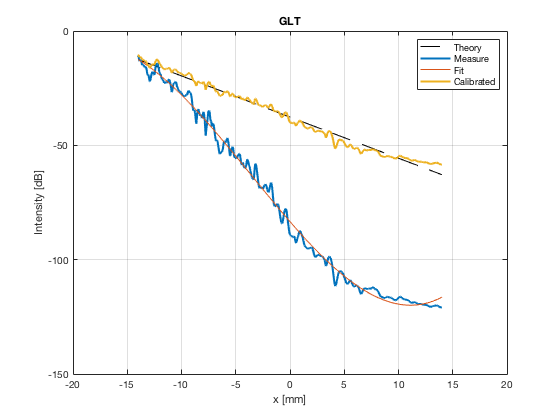
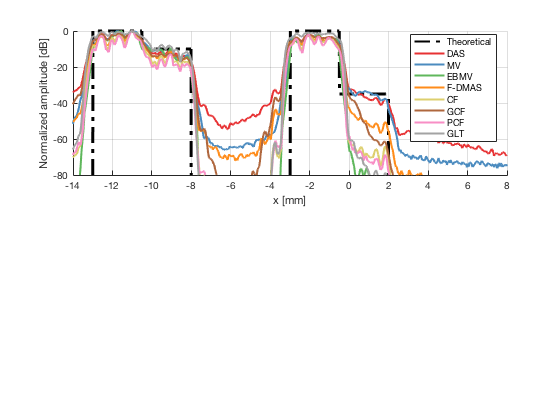
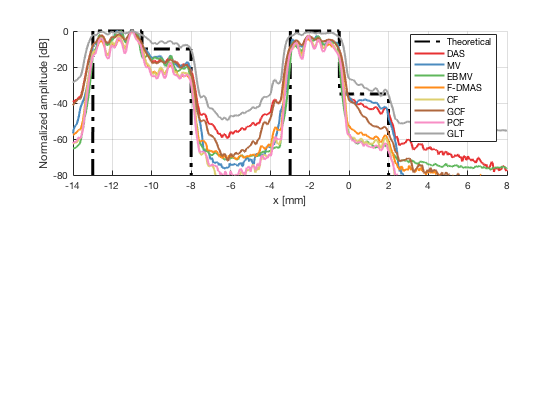
Plot the lateral line through the boxes after calibration
[meanLines,x_axis] = getMeanLateralLines(b_data_das,image_calibrated_on_theoretical,27.5,32.5,b_data_das.scan.x(1)*10^3,b_data_das.scan.x(end)*10^3); theoretical = [-100 ones(1,255)*0 ones(1,255)*-10 -100 ones(1,256)*-100 ones(1,256)*-100 -100 ones(1,255)*0 ones(1,255)*-35 -100]; x_axis_theoretical = linspace(-13,2,1536); f89 = figure(90);clf;hold all; subplot(211);hold on; plot(x_axis_theoretical,theoretical,'LineStyle','-.','Linewidth',2,'DisplayName','Theoretical','Color',[0 0 0]); plot(x_axis,meanLines.all{1}-max(meanLines.all{1}),'Linewidth',2,'DisplayName',image.tags{1},'Color',colors(1,:)); plot(x_axis,meanLines.all{2}-max(meanLines.all{2}),'Linewidth',2,'DisplayName',image.tags{2},'Color',colors(2,:)); plot(x_axis,meanLines.all{3}-max(meanLines.all{3}),'Linewidth',2,'DisplayName',image.tags{3},'Color',colors(3,:)); plot(x_axis,meanLines.all{4}-max(meanLines.all{4}),'Linewidth',2,'DisplayName',image.tags{4},'Color',colors(4,:)); plot(x_axis,meanLines.all{5}-max(meanLines.all{5}),'Linewidth',2,'DisplayName',image.tags{5},'Color',colors(5,:)); plot(x_axis,meanLines.all{6}-max(meanLines.all{6}),'Linewidth',2,'DisplayName',image.tags{6},'Color',colors(6,:)); plot(x_axis,meanLines.all{7}-max(meanLines.all{7}),'Linewidth',2,'DisplayName',image.tags{7},'Color',colors(7,:)); plot(x_axis,meanLines.all{8}-max(meanLines.all{8}),'Linewidth',2,'DisplayName',image.tags{8},'Color',colors(8,:)); ylim([-80 0]); xlim([-14 8]); legend('Location','best'); grid on; ylabel('Normalized amplitude [dB]'); xlabel('x [mm]');
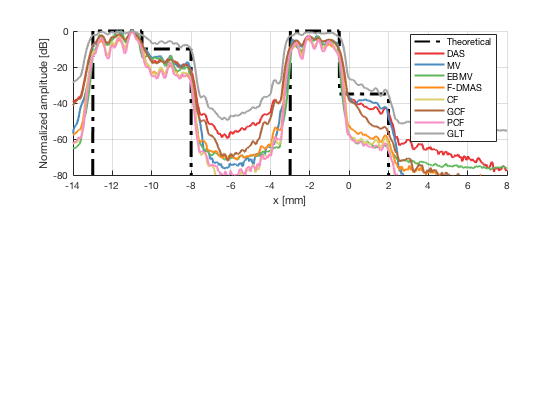
saveas(f89,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/calibrated/boxes_calibrated_lines_on_theoretical'],'eps2c') saveas(f89,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/calibrated/boxes_calibrated_lines_on_theoretical'],'png')
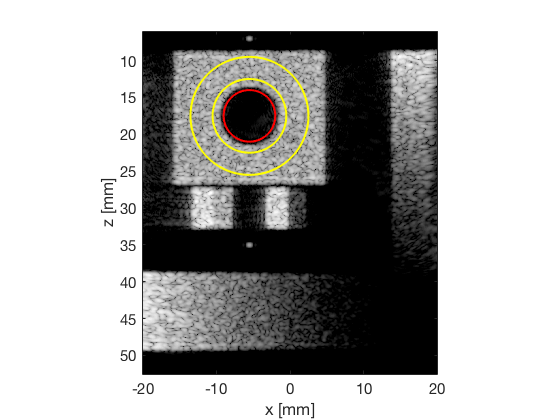
Measure contrast and plot together with the experimental
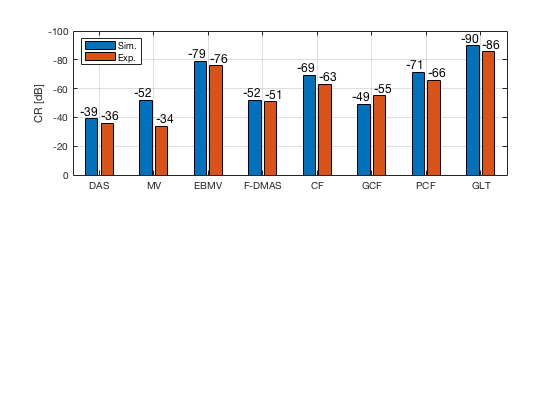
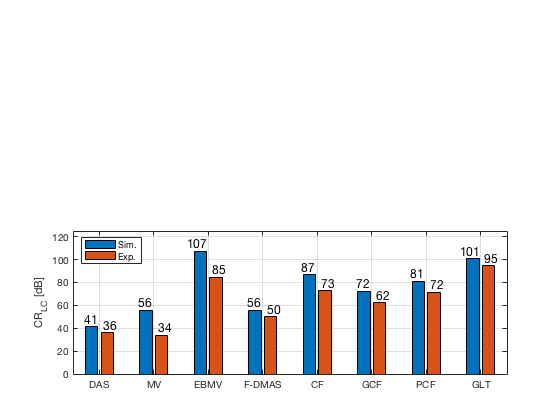
[CR_signal_sim, CR_signal_dagger, CR_image_sim, CNR_signal_sim, CNR_image_sim] = measureContrast(b_data_das,image,-5.5,17.5,3.5,5,8,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/DAS_cyst_indicated']); load('Experimental.mat','CR_signal_exp','CNR_signal_exp','CR_image_exp','CNR_image_exp');
f9 = figure; subplot(211); bar([10*log10(CR_signal_sim)' 10*log10(CR_signal_exp)']) set(gca,'XTick',linspace(1,length(image.tags),length(image.tags))) set(gca,'XTickLabel',image.tags) ylabel('CR [dB]'); x_pos = [0.8 1.8 2.8 3.8 4.8 5.8 6.8 7.8]; text(x_pos,double(10*log10(CR_signal_sim(:))),num2str(round(10*log10(CR_signal_sim(:))),'%d'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) x_pos = [1.2 2.2 3.2 4.2 5.2 6.2 7.2 8.2]; text(x_pos,double(10*log10(CR_signal_exp(:))),num2str(round(10*log10(CR_signal_exp(:))),'%d'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) set(gca,'Ydir','reverse') legend('Sim.','Exp.','Location','nw') grid on
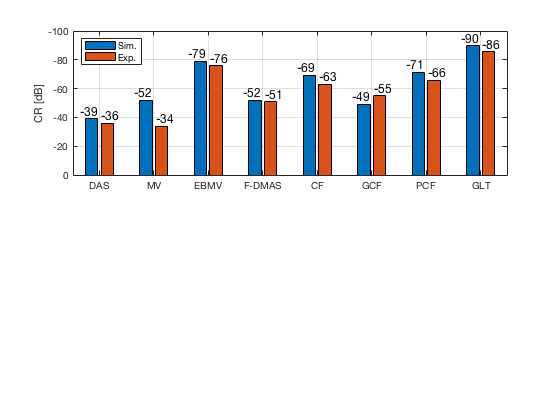
saveas(f9,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/CR'],'eps2c')
f90 = figure; subplot(212); bar([CR_image_sim' CR_image_exp']) set(gca,'XTick',linspace(1,length(image.tags),length(image.tags))) set(gca,'XTickLabel',image.tags) ylabel('CR_{LC} [dB]'); x_pos = [0.8 1.8 2.8 3.8 4.8 5.8 6.8 7.8]; text(x_pos,double(CR_image_sim(:)),num2str(round(CR_image_sim(:)),'%d'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) x_pos = [1.2 2.2 3.2 4.2 5.2 6.2 7.2 8.2]; text(x_pos,double(CR_image_exp(:)),num2str(round(CR_image_exp(:)),'%d'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) ylim([0 125]) legend('Sim.','Exp.','Location','nw') grid on
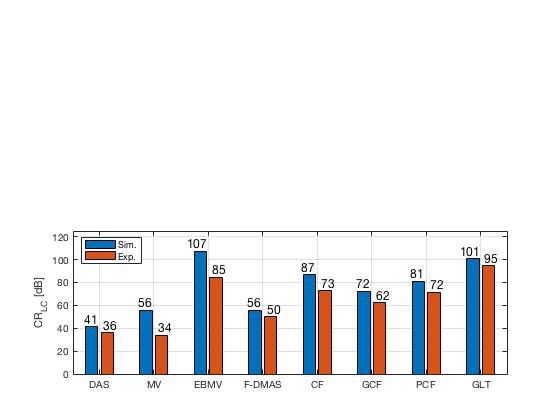
saveas(f90,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/CR_LC'],'eps2c')
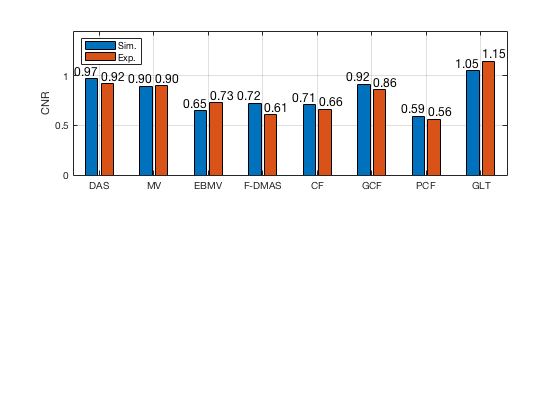
f77 = figure(77);clf; subplot(211); bar([CNR_signal_sim' CNR_signal_exp']) set(gca,'XTick',linspace(1,length(image.tags),length(image.tags))) set(gca,'XTickLabel',image.tags) ylabel('CNR'); x_pos = [0.75 1.75 2.75 3.75 4.75 5.75 6.75 7.75]; text(x_pos,double(CNR_signal_sim(:)),num2str((CNR_signal_sim(:)),'%.2f'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) x_pos = [1.25 2.25 3.25 4.25 5.25 6.25 7.25 8.25]; text(x_pos,double(CNR_signal_exp(:)),num2str((CNR_signal_exp(:)),'%.2f'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) ylim([0 1.45]) legend('Sim.','Exp.','Location','nw') grid on
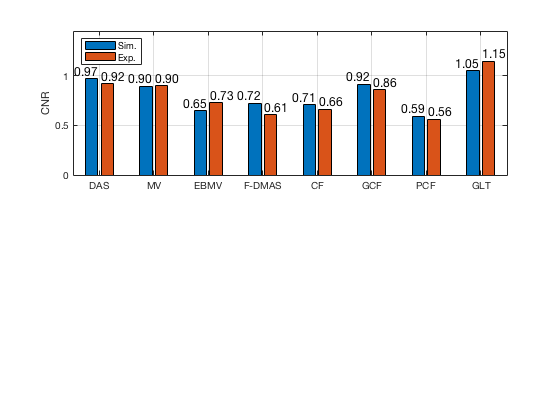
saveas(f77,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/CNR'],'eps2c')
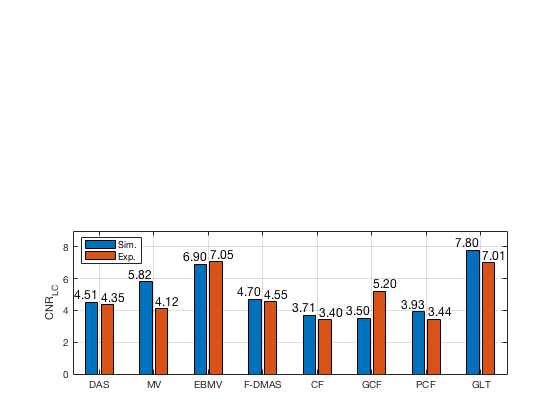
f78 = figure(78);clf; subplot(212); bar([CNR_image_sim' CNR_image_exp']) set(gca,'XTick',linspace(1,length(image.tags),length(image.tags))) set(gca,'XTickLabel',image.tags) ylabel('CNR_{LC}'); x_pos = [0.75 1.75 2.75 3.75 4.75 5.75 6.75 7.75]; text(x_pos,double(CNR_image_sim(:)),num2str((CNR_image_sim(:)),'%.2f'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) x_pos = [1.25 2.25 3.25 4.25 5.25 6.25 7.25 8.25]; text(x_pos,double(CNR_image_exp(:)),num2str((CNR_image_exp(:)),'%.2f'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) legend('Sim.','Exp.','Location','nw') grid on ylim([0 9])
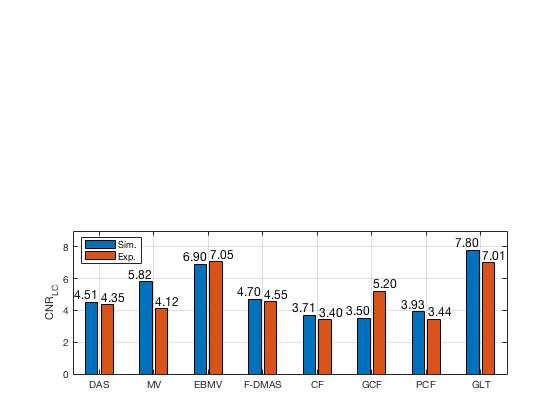
saveas(f78,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/CNR_LC'],'eps2c')
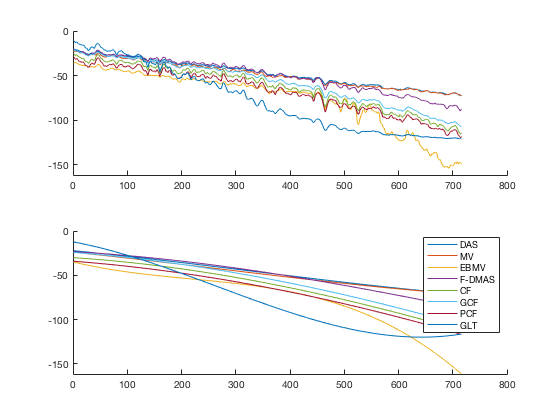
Run Dynamic Range Test
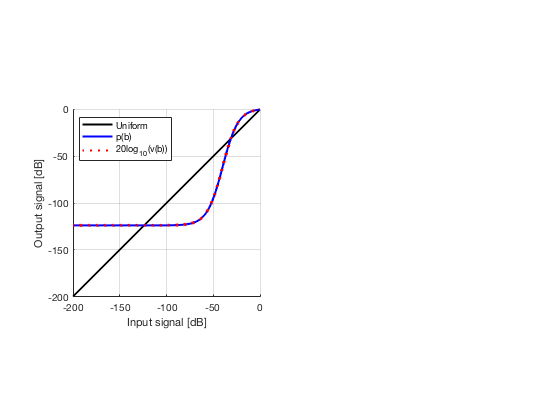
diff{1} = tools.dynamic_range_test(channel_data,b_data_das,'DAS');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','DAS'],'eps2c');
diff{2} = tools.dynamic_range_test(channel_data,b_data_mv,'MV');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','MV'],'eps2c');
diff{3} = tools.dynamic_range_test(channel_data,b_data_ebmv,'EBMV');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','EBMV'],'eps2c');
diff{4} = tools.dynamic_range_test(channel_data,b_data_dmas,'F-DMAS');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','F-DMAS'],'eps2c');
diff{5} = tools.dynamic_range_test(channel_data,b_data_cf,'CF');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','CF'],'eps2c');
diff{6} = tools.dynamic_range_test(channel_data,b_data_gcf,'GCF');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','GCF'],'eps2c');
diff{7} = tools.dynamic_range_test(channel_data,b_data_pcf,'PCF');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','PCF'],'eps2c');
diff{8} = tools.dynamic_range_test(channel_data,b_data_glt,'GLT');
set(gcf,'Position',[7 222 1271 347]);
saveas(gcf,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/dynamic_range_test/','GLT'],'eps2c');
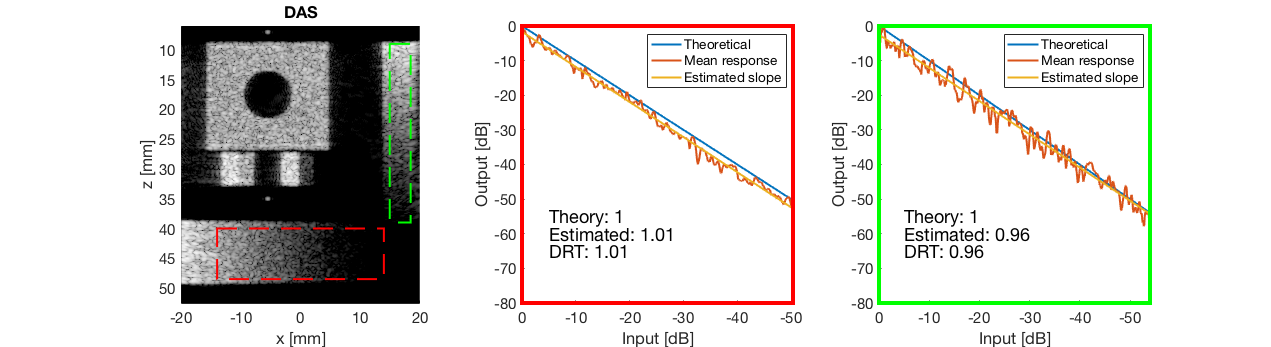
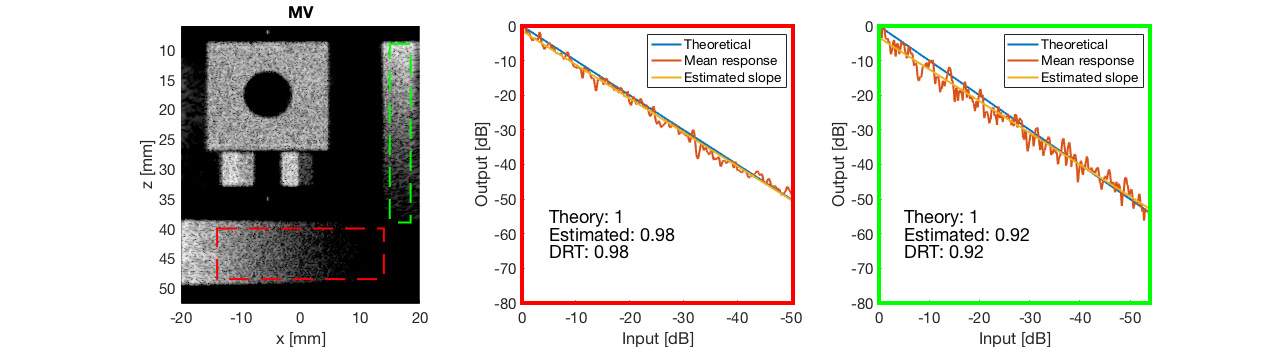
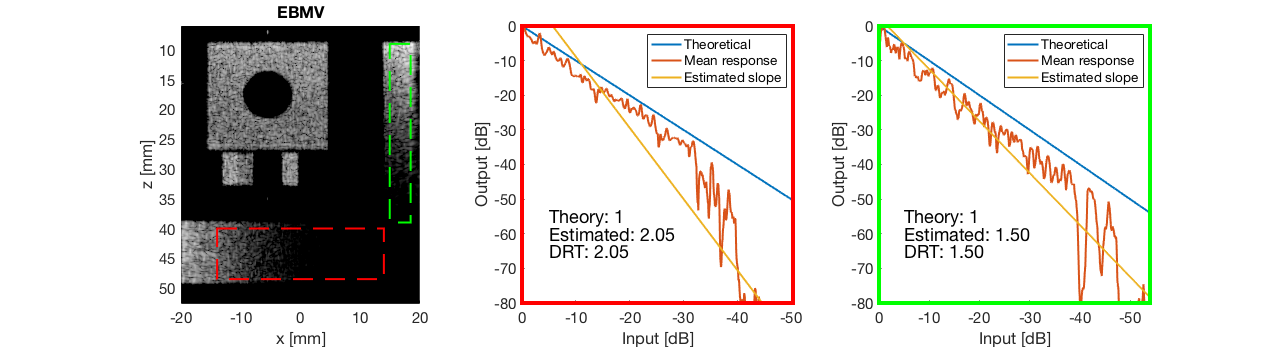
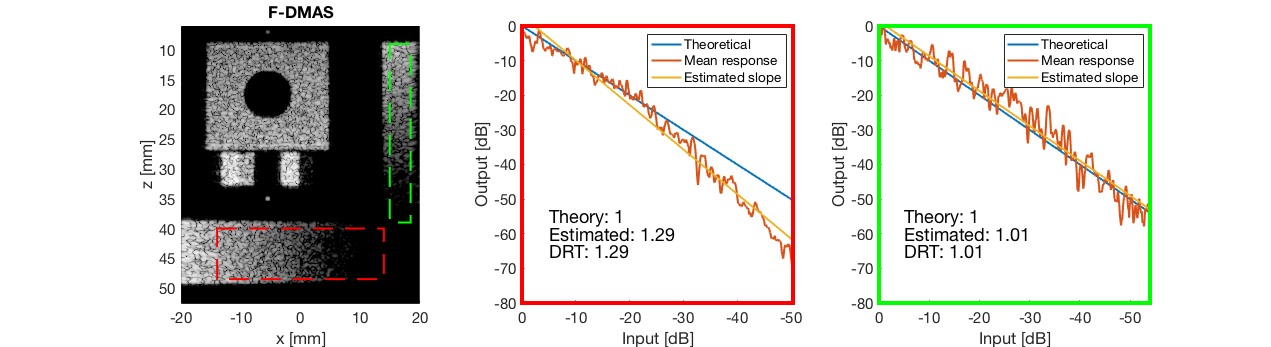
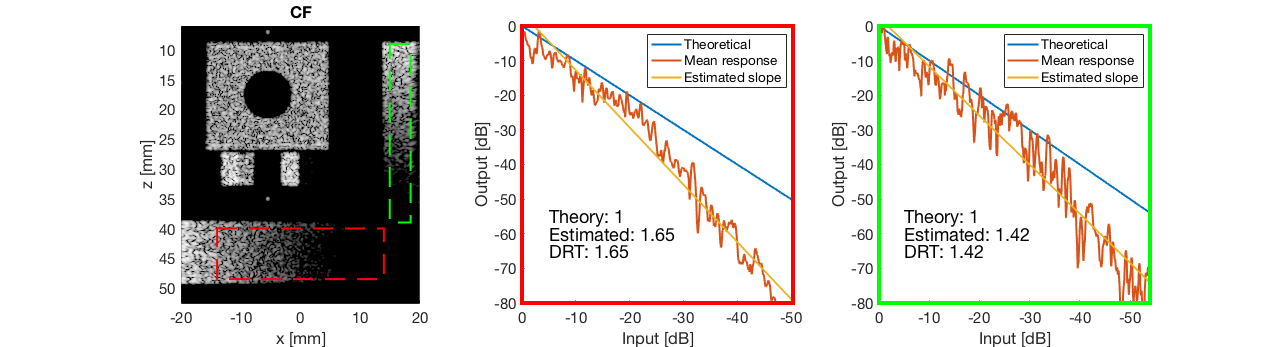
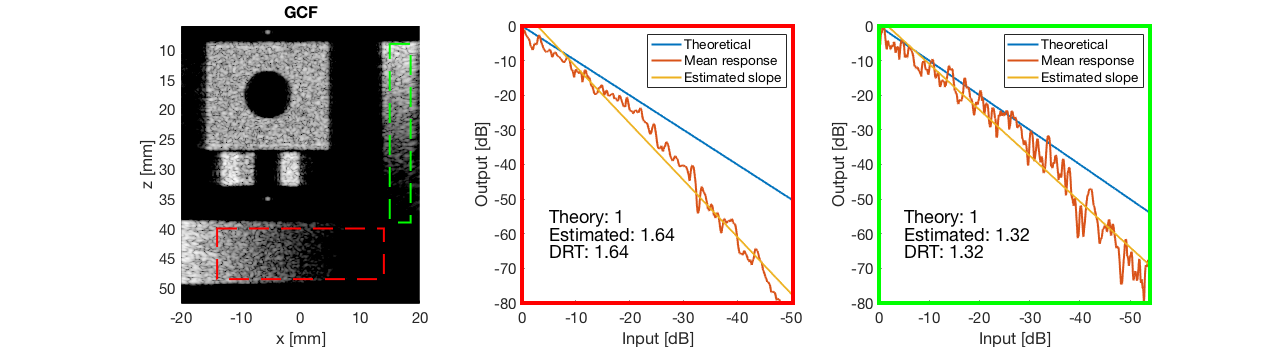
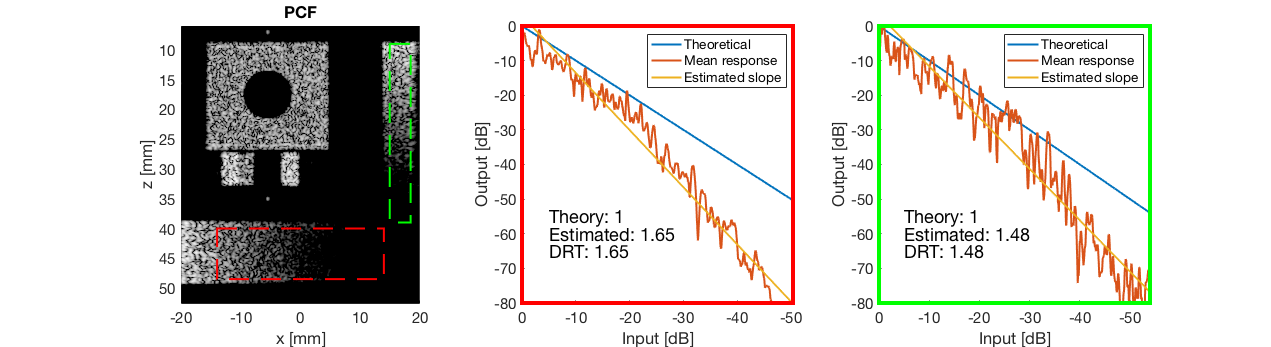
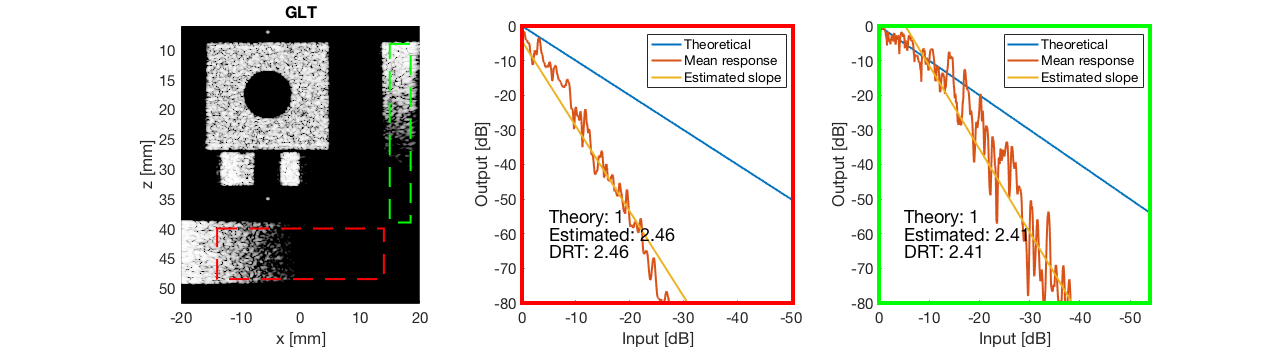
for i = 1:length(image.tags) DRT_value(i) = (mean(diff{i})); end
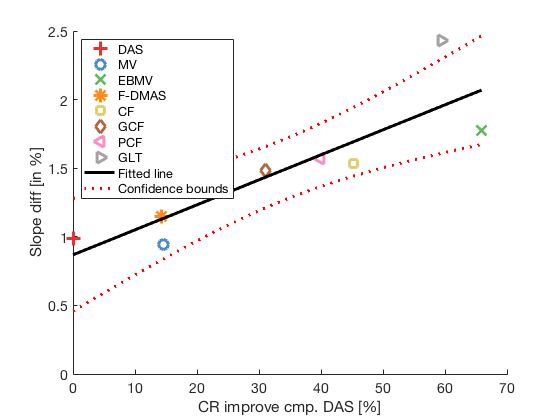
cr_improvement = CR_image_sim - CR_image_sim(1); lmd_sim_slope = fitlm(cr_improvement(1:8),DRT_value(1:8)) lmd_sim_slope.Rsquared.Ordinary lmd_sim_slope.Rsquared.Adjusted f113 = figure(114);clf;hold all; linespes = '+ox*sd<>'; for i = 1:length(image.all) plot(cr_improvement(i),DRT_value(i),linespes(i),'Color',colors(i,:),'MarkerSize',10); end lmd_sim_slope.plot hline = findobj(gcf, 'type', 'line'); set(hline,'LineWidth',3); set(hline(3),'Color',[0 0 0]); set(hline(3),'DisplayName','Fitted line'); title('') xlabel('CR improve cmp. DAS [%]');%xlim([-5 75]); ylabel('Slope diff [in %]');%ylim([-50 5]); text(80,10,sprintf('R-Squared: %.2f',lmd_sim_slope.Rsquared.Ordinary),'FontSize',18); text(80,0,sprintf('R-Squared adj: %.2f',lmd_sim_slope.Rsquared.Adjusted),'FontSize',18); set(gca,'FontSize',14); delete(hline(4)); legend(); hLegend = findobj(gcf, 'Type', 'Legend'); legend show legend([{image.tags{1:8}} hLegend.String],'Location','nw'); saveas(f113,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/correlation_simulation'],'eps2c')
lmd_sim_slope =
Linear regression model:
y ~ 1 + x1
Estimated Coefficients:
Estimate SE tStat pValue
________ ________ ______ _________
(Intercept) 0.86838 0.16773 5.1773 0.0020598
x1 0.018225 0.004179 4.361 0.0047651
Number of observations: 8, Error degrees of freedom: 6
Root Mean Squared Error: 0.256
R-squared: 0.76, Adjusted R-Squared 0.72
F-statistic vs. constant model: 19, p-value = 0.00477
ans =
0.7602
ans =
0.7202
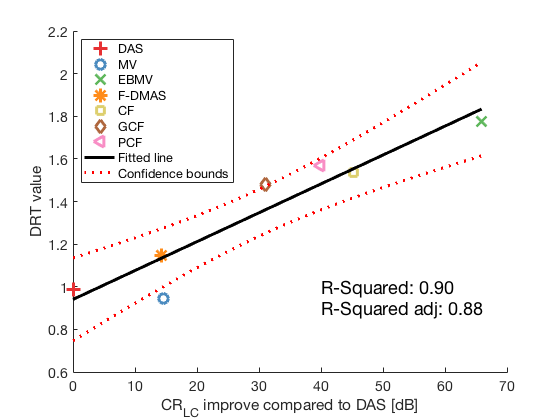
lmd_sim_slope = fitlm(cr_improvement(1:7),DRT_value(1:7)) lmd_sim_slope.Rsquared.Ordinary lmd_sim_slope.Rsquared.Adjusted f113 = figure(114);clf;hold all; linespes = '+ox*sd<>'; for i = 1:length(image.all)-1 plot(cr_improvement(i),DRT_value(i),linespes(i),'Color',colors(i,:),'MarkerSize',10); end lmd_sim_slope.plot hline = findobj(gcf, 'type', 'line'); set(hline,'LineWidth',3); set(hline(3),'Color',[0 0 0]); set(hline(3),'DisplayName','Fitted line'); title('') xlabel('CR_{LC} improve compared to DAS [dB]','Interpreter', 'tex'); ylabel('DRT value'); text(40,1,sprintf('R-Squared: %.2f',lmd_sim_slope.Rsquared.Ordinary),'FontSize',18); text(40,0.9,sprintf('R-Squared adj: %.2f',lmd_sim_slope.Rsquared.Adjusted),'FontSize',18); set(gca,'FontSize',14); delete(hline(4)); legend(); hLegend = findobj(gcf, 'Type', 'Legend'); legend show legend([{image.tags{1:7}} hLegend.String],'Location','nw'); saveas(f113,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/correlation_simulation_without_glt'],'eps2c')
lmd_sim_slope =
Linear regression model:
y ~ 1 + x1
Estimated Coefficients:
Estimate SE tStat pValue
________ _________ ______ __________
(Intercept) 0.94049 0.075572 12.445 5.9395e-05
x1 0.013554 0.0020665 6.5588 0.001235
Number of observations: 7, Error degrees of freedom: 5
Root Mean Squared Error: 0.113
R-squared: 0.896, Adjusted R-Squared 0.875
F-statistic vs. constant model: 43, p-value = 0.00123
ans =
0.8959
ans =
0.8750
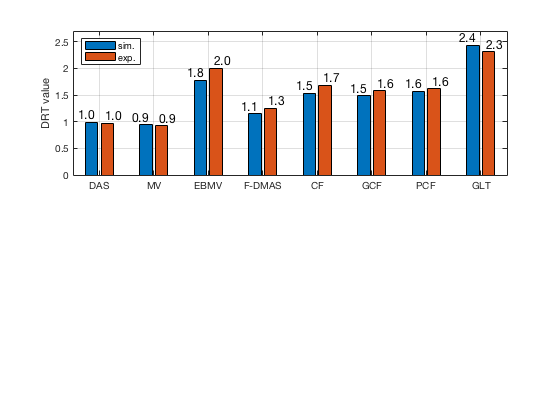
load([ustb_path,'/publications/DynamicRange/','Experimental.mat'],'DRT_value_exp'); f10 = figure(10);clf; subplot(211); bar([DRT_value' DRT_value_exp']) set(gca,'XTick',linspace(1,length(image.tags),length(image.tags))) set(gca,'XTickLabel',image.tags) ylabel('DRT value'); legend('Location','nw','sim.','exp.') x_pos = [0.75 1.25 1.75 2.25 2.75 3.25 3.75 4.25 4.75 5.25 5.75 6.25 6.75 7.25 7.75 8.25]; DRT_value_all = []; for i = 1:length(DRT_value_exp) DRT_value_all = [DRT_value_all; DRT_value(i); DRT_value_exp(i)]; end text(x_pos,double(DRT_value_all),num2str(DRT_value_all(:),'%0.1f'),... 'HorizontalAlignment','center',... 'VerticalAlignment','bottom',... 'FontSize',12) grid on; ylim([0 2.7]) saveas(f10,[ustb_path,filesep,'publications/DynamicRange/figures/simulation/DRT'],'eps2c')