Reading data from an UFF file
In this example we show how to read channel and beamformed data from a UFF (Ultrasound File Format) file. You will need an internet connection to download data. Otherwise, you can run the CPWC_UFF_write.m first so the file 'test02.uff' is in the current path.
by Alfonso Rodriguez-Molares alfonso.r.molares@ntnu.no 15.05.2017
Contents
Checking the file is in the path
To read data from a UFF file the first we need is, you guessed it, a UFF file. We check if it is on the current path and download it from the NTNU server otherwise.
% data location url='http://hirse.medisin.ntnu.no/ustb/data/uff/'; % if not found data will be downloaded from here local_path = [ustb_path(),'/data/']; % location of example data in this computer addpath(local_path); filename='test02.uff'; % check if the file is available in the local path & downloads otherwise tools.download(filename, url, local_path);
Reading beamformed data
Now that the file is on the path let us create a UFF object to interact with the file.
uff_file=uff(filename,'read');
We don't want to screw up the file so we open it in "read-only" mode. Now we can have a peek at what is inside with the index method
display=true;
uff_file.index('/',display);
UFF: Contents of test02.uff at / - /b_data: b_data [uff.beamformed_data] size(1,1) - /b_data_array: b_data_array [uff.beamformed_data] size(1,3) - /b_data_copy: b_data_copy [uff.beamformed_data] size(1,1) - /channel_data: channel_data [uff.channel_data] size(1,1) - /scan: scan [uff.linear_scan] size(1,1)
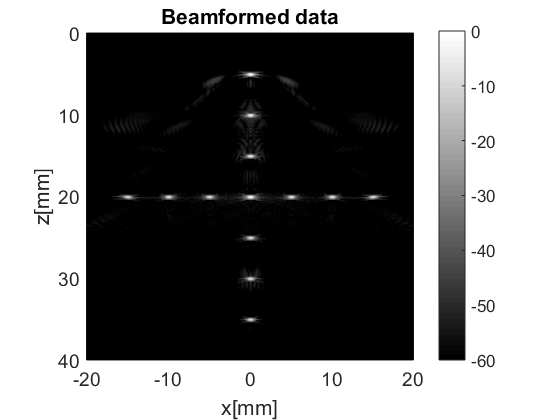
We see there is a beamformed_data dataset with name b_data. Let us load it and plot it.
b_data=uff_file.read('/b_data');
b_data.plot();
UFF: reading b_data [uff.beamformed_data]
There is also an array of beamformed images with name b_data_array. Let us load it too
b_data_array=uff_file.read('/b_data_array'); figure; for n=1:length(b_data_array) b_data.plot(subplot(1,length(b_data_array),n)); end set(gcf,'Position',[0 0 1000 420])
UFF: reading b_data_array [uff.beamformed_data] UFF: reading b_data_array [uff.beamformed_data] UFF: reading b_data_array_0001 [uff.beamformed_data] Processed 1/3UFF: reading b_data_array_0002 [uff.beamProcessed 2/3UFF: reading b_data_array_0003 [uff.beamProcessed 3/3
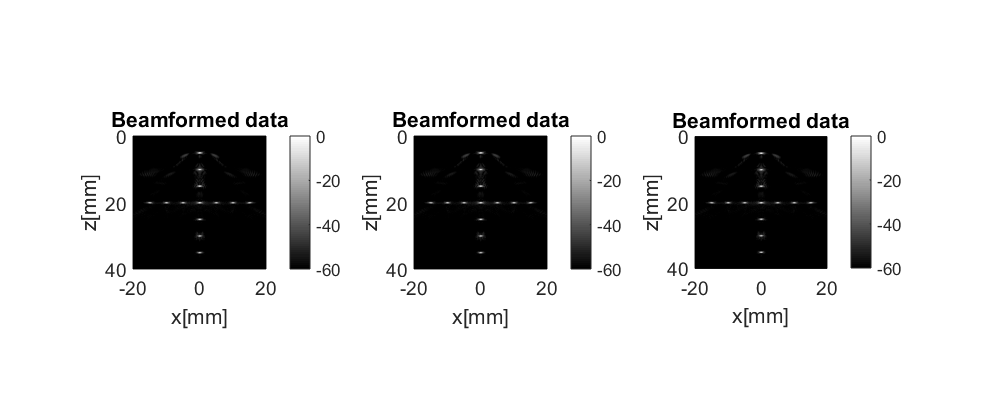
Not very interesting, actually.
Reading channel data
There are also two other structures in the file: a uff.scan and a uff.channel_data objects. Let us read them both
scan=uff_file.read('/scan'); channel_data=uff_file.read('/channel_data');
UFF: reading channel_data [uff.channel_data] UFF: reading phantom [uff.phantom] UFF: reading sequence [uff.wave] Processed 31/31
And let us beamform that data with USTB
bmf=beamformer();
bmf.channel_data=channel_data;
bmf.scan=scan;
bmf.receive_apodization.window=uff.window.tukey50;
bmf.receive_apodization.f_number=1.0;
bmf.receive_apodization.apex.distance=Inf;
bmf.transmit_apodization.window=uff.window.tukey50;
bmf.transmit_apodization.f_number=1.0;
bmf.transmit_apodization.apex.distance=Inf;
% beamforming
b_data=bmf.go({process.das_mex() process.coherent_compounding()});
b_data.plot();
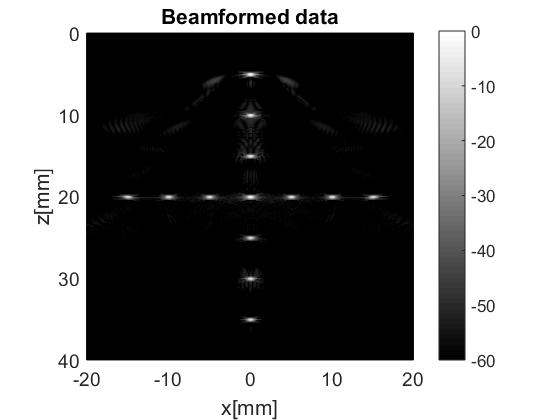
which matches the images we saw previously.
Reading once and for all
It is possible to load all the data in the file into matlab memory without having to access each dataset individually. It suffices to call the read method without parameters and...
vars=uff_file.read();
UFF: reading b_data [uff.beamformed_data] UFF: reading b_data_array [uff.beamformed_data] UFF: reading b_data_array [uff.beamformed_data] UFF: reading b_data_array_0001 [uff.beamformed_data] Processed 1/3UFF: reading b_data_array_0002 [uff.beamProcessed 2/3UFF: reading b_data_array_0003 [uff.beamProcessed 3/3 UFF: reading b_data_copy [uff.beamformed_data] UFF: reading channel_data [uff.channel_data] UFF: reading phantom [uff.phantom] UFF: reading sequence [uff.wave] Processed 31/31
... we get all the objects in the file into a cell structure
vars{1}.plot();