Delay Multiply And Sum on FI data from an UFF file
This script is also available as /publications/TUFFC/Prieur_et_al_Signal_coherence_and_image_amplitude_with_the_fDMAS /FI_UFF_delay_multiply_and_sum_Fig5_and_Fig6.m in the USTB repository.
Create the images with the recorded channel data in the paper:
F. Prieur, O. M. H. Rindal and A. Austeng, "Signal coherence and image amplitude with the Filtered-Delay-Multiply-And-Sum beamformer," in IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. doi: 10.1109/TUFFC.2018.2831789
This script re-create Figs. 5 and 6 of the article.
This code uses the UltraSound ToolBox (USTB) and you'll need to download it first to successfully rund this code. To know more about the USTB visit http://www.ustb.no/
by Ole Marius Hoel Rindal olemarius@olemarius.net 10.10.2017
Contents
- Setting up file path
- Reading channel data from UFF file
- Define Scan
- Delay the channel data
- Define the DAS beamformer using coherent compounding of delayed data
- Create the DMAS image using the delay_multiply_and_sum postprocess
- Calculate the coherence factor (CF) image
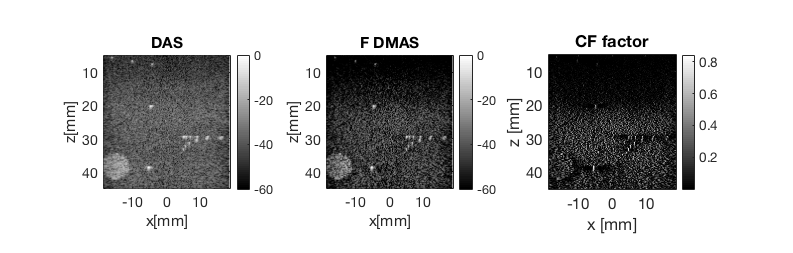
- Plot all three images in same plot - Fig. 5
Setting up file path
To read data from a UFF file the first we need is, you guessed it, a UFF file. We check if it is on the current path and download it from the USTB websever.
clear all; close all; % data location url='http://ustb.no/datasets/'; % if not found downloaded from here local_path = [ustb_path(),'/data/']; % location of example data % Choose dataset filename='L7_FI_Verasonics_CIRS_points.uff'; % check if the file is available in the local path or downloads otherwise tools.download(filename, url, local_path);
Reading channel data from UFF file
channel_data=uff.read_object([local_path filename],'/channel_data');
UFF: reading /channel_data [uff.channel_data] UFF: reading sequence [uff.wave] [====================] 100%
%Print info about the dataset
channel_data.print_authorship
Name: Dataset recorded on a Verasonics Vantage 256. Focused Imaging using L7-4 linear probe Reference: F. Prieur, O. M. H. Rindal and A. Austeng, "Signal coherence and image amplitude with the Filtered-Delay-Multiply-And-Sum beamformer," in IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control. doi: 10.1109/TUFFC.2018.2831789 Author(s): Ole Marius Hoel Rindal <olemarius@olemarius.net> Fabruce Prieur <fabrice@ifi.uio.no> Version:
Define Scan
Define the image coordinates we want to beamform in the scan object. Notice that we need to use quite a lot of samples in the z-direction. This is because the DMAS creates an "artificial" second harmonic signal, so we need high enough sampling frequency in the image to get a second harmonic signal.
z_axis=linspace(5e-3,45e-3,1700).'; x_axis=zeros(channel_data.N_waves,1); for n=1:channel_data.N_waves x_axis(n) = channel_data.sequence(n).source.x; end scan=uff.linear_scan('x_axis',x_axis,'z_axis',z_axis);
Delay the channel data
delay = midprocess.das(); delay.channel_data=channel_data; delay.scan=scan; delay.dimension = dimension.transmit(); delay.transmit_apodization.window=uff.window.scanline; delay.receive_apodization.window=uff.window.none; delay.receive_apodization.f_number=1.7; delayed_b_data = delay.go();
USTB General beamformer MEX v1.1.2 .............done!
Define the DAS beamformer using coherent compounding of delayed data
das = postprocess.coherent_compounding();
das.input = delayed_b_data;
b_data_das = das.go();
b_data_das.plot([],'DAS');
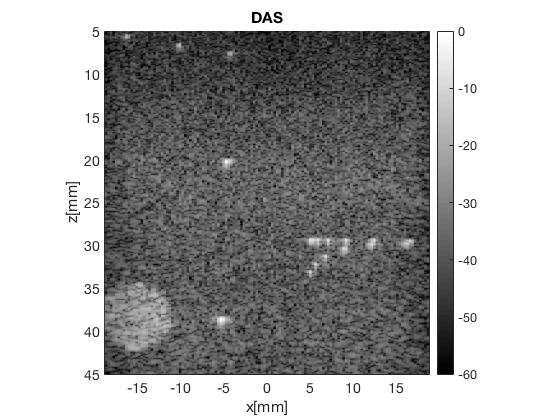
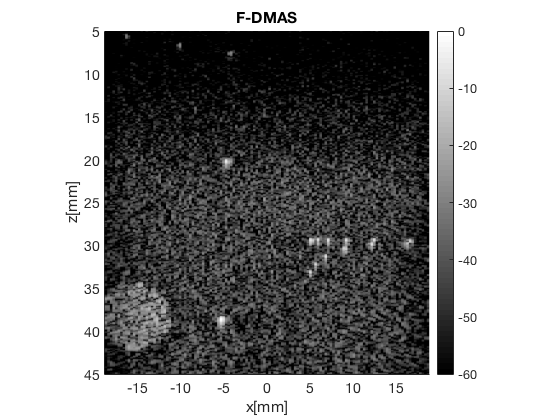
Create the DMAS image using the delay_multiply_and_sum postprocess
dmas = postprocess.delay_multiply_and_sum(); dmas.dimension = dimension.receive; dmas.channel_data = channel_data; dmas.input = delayed_b_data; dmas.receive_apodization = delay.receive_apodization; b_data_dmas=dmas.go(); % beamforming b_data_dmas.plot(100,'F-DMAS');
Warning: Missing probe and apodization data; full aperture is assumed. Warning: If the result looks funky, you might need to tune the filter paramters of DMAS using the filter_freqs property. Use the plot to check that everything is OK.
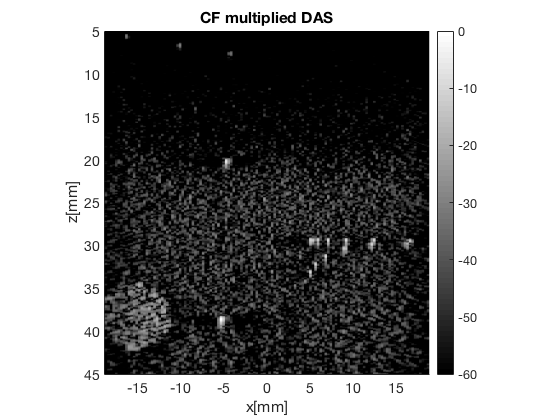
Calculate the coherence factor (CF) image
cf = postprocess.coherence_factor(); cf.dimension = dimension.receive(); cf.input = delayed_b_data; b_data_CF_multiplied_DAS = cf.go(); b_data_CF_multiplied_DAS.plot(101,'CF multiplied DAS'); % Get just the CF "factor" cfImg = cf.CF.get_image('none');
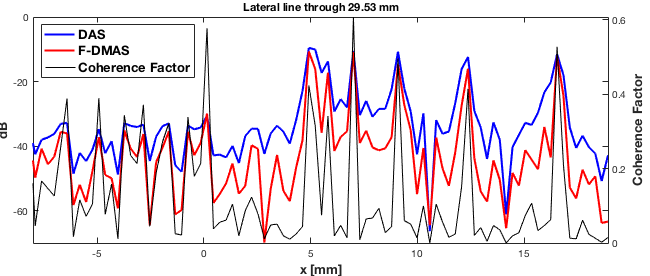
Plot all three images in same plot - Fig. 5
f3 = figure(3);clf b_data_das.plot(subplot(1,3,1),'DAS'); % Display image ax(1) = gca; b_data_dmas.plot(subplot(1,3,2),'F DMAS'); % Display image ax(2) = gca; subplot(1,3,3) imagesc(scan.x_axis*1e3,scan.z_axis*1e3,cfImg);colormap gray;title('CF factor'); xlabel('x [mm]');ylabel('z [mm]'); colorbar;axis('image') set(gca,'FontSize',15); ax(3) = gca; linkaxes(ax); set(gcf,'Position',[195 446 793 253]);
dmas_img = b_data_dmas.get_image(); das_img = b_data_das.get_image(); line_idx = find(b_data_dmas.scan.z_axis>=29.52e-3,1,'first'); x_axis=b_data_dmas.scan.x_axis; pdmas=dmas_img(line_idx,:); pdas=das_img(line_idx,:); cfl=cfImg(line_idx,:); % Plotting Fig. 6 figure('color','w','position',[244 164 650 276]); subplot('position',[0.05 0.12 0.885 0.82]) [hAx,hLine1,hLine2] = plotyy([x_axis*1000, x_axis*1000],[pdas.', pdmas.'],x_axis*1000,cfl); hLine1(1).LineWidth=2;hLine1(1).Color='b'; hLine1(2).LineWidth=2;hLine1(2).Color='r'; hLine2.Color='k';%hLine2.LineStyle='--'; hAx(1).YLim=[-70 0];hAx(1).XLim=[-8 max(x_axis*1000)]; hAx(2).YLim=[0 max(cfl)];hAx(2).XLim=[-8 max(x_axis*1000)]; hAx(1).YLabel.String='dB';hAx(1).YLabel.FontWeight='bold'; hAx(2).YLabel.String='Coherence Factor';hAx(2).YLabel.FontWeight='bold'; hAx(1).YLabel.FontSize=13;hAx(2).YLabel.FontSize=13; hAx(1).XLabel.String='x [mm]';hAx(1).XLabel.FontWeight='bold'; hAx(1).XLabel.FontSize=13; hAx(2).YColor=hAx(1).YColor;grid title(sprintf('Lateral line through %.2f mm',... b_data_dmas.scan.z_axis(line_idx)*10^3)); h=legend('DAS','F-DMAS','Coherence Factor','location','northwest');grid set(h,'fontsize',13,'fontweight','bold')